Staphylococcus aureus functional amyloids catalyze degradation of β-lactam antibiotics
Published in Chemistry and Microbiology
Antibiotic resistance by pathogenic bacteria is among the most pressing challenges in modern medicine and healthcare1. Antibiotic resistance is particularly acute in the case of β-lactams, among the most widely used class of antibiotics, which include penicillin, penems, and others2. This study demonstrates that bacterial functional amyloids catalyze degradation of β-lactams, illustrating a possible new pathway for antibiotic resistance by bacteria. Amyloid fibrils constitute important components of bacterial biofilms, specifically comprising biofilm scaffolding3. Amyloids of Staphylococcus aureus belonging to the phenol-soluble modulin alpha (PSMα) family, which are the focus of this work, have been shown to form unique cross-α organizations4,5.
Amyloidogenic proteins have been shown to exhibit catalytic properties6. While almost all previous studies have focused on synthetic de-novo peptides, we have recently demonstrated that native, physiological amyloids can catalyze disease-related and key metabolic reactions7,8. The present study reveals that PSMα amyloids, particularly PSMα3, catalyse hydrolysis of a prominent amide-like bond in β-lactam antibiotics, thereby rendering the molecules inactive. Through structure/function analyses of PSMα3 variants, putative catalytic sites on the amyloid fibrils’ surface were localized, linked to both the cross-α organization of the amyloid fibrils and electrostatic interactions between the β-lactam molecules and lysine sidechains. Amyloid-catalyzed β-lactam degradation may underscore an intriguing new paradigm accounting for antibiotic resistance of S. aureus, and pathogenic bacteria in general.
The hypothesis and experimental scheme underlining this study are illustrated in Figure 1a. PSMα amyloid fibrils (physiologically identified as constituents of S. auereus biofilms, and also shown to be secreted by this bacterial species as toxins9,10) adsorb β-lactams which are subsequently degraded through catalyzing hydrolysis of the 4-member β-lactam ring. The experiments presented in the manuscript were designed to examine whether PSMα amyloids, through active sites upon the amyloid fibril surface (schematically represented as “scissors”), catalyze cleavage of the four-membered β-lactam ring, thereby making the molecule functionally inactive. Specifically, we tested PSMα amyloid-mediated hydrolysis of nitrocefin, a widely-studied β-lactam surrogate (Figure 1b)11.
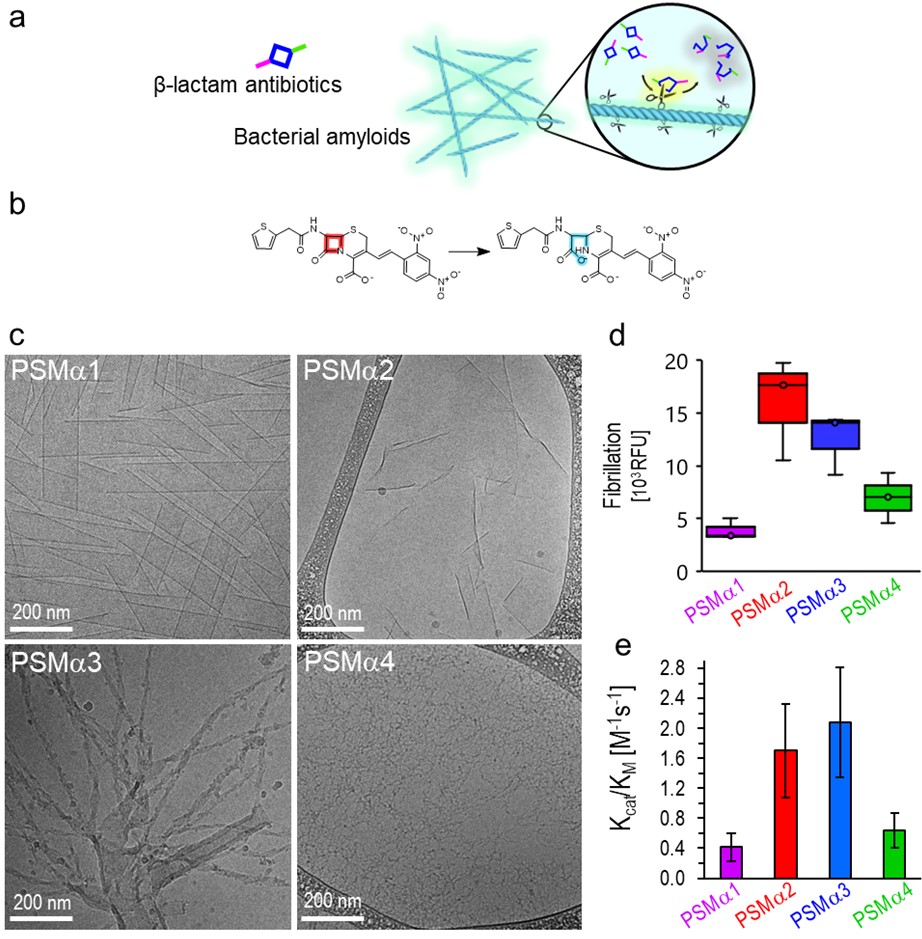
Figure 1. S. aureus-secreted functional amyloids: Hypothesis and structural characterization of the PSMα amyloids. a. PSMα amyloid fibrils bind β-lactams and catalyze their degradation through amide bond hydrolysis. b. Nitrocefin degradation via the four-member ring opening – a model reaction for β-lactam degradation. c. Cryo-TEM images of the PSMα1-4 amyloids. d. Quantification of amyloid formation using AmytrackerTM fluorescent labelling. e. The catalytic efficiencies (Kcat/KM) of PSMα1-4 amyloids in the case of nitrocefin degradation. This measure, based on kinetic experiments of nitrocefin degradation, shows that PSMα2 and PSMα3 amyloids were the most catalytically active.
Figure 1c-d present characterization of the PSMα1-4 amyloid fibrils investigated: The cryogenic transmission electron microscopy (cryo-TEM) images in Figure 1c illuminate the amyloid morphologies of the peptides. Quantification of PSMα fibrillar samples incubated with the amyloid-specific dye Amytracker-680, which fluorescence intensity correlates with repeating amyloidogenic domains12,13, are portrayed in Figure 1d, indicating variability in the extent of amyloid formation by the PSMα peptides. Specifically, PSMα2 and PSMα3 gave rise to high fluorescence of the dye, indicating pronounced amyloid organization, while PSMα1 and PSMα4 generated lower Amytracker-680 fluorescence, reflecting lesser amyloid fibril organization of the peptide aggregates.
Figure 1e depicts PSMα-mediated catalysis of β-lactam ring breakup via amide bond hydrolysis, which correlated with the amyloid abundance data in Figure 1d. In the experiments, we recorded the degradation kinetics of nitrocefin following addition of the PSMα amyloids and calculated the catalytic efficiencies, (Kcat/KM), through fitting the kinetic curves to the Michaelis-Menten (MM) enzymatic model. Consistent with the structural data, there is a strong correlation between amyloid formation and abundance and catalytic activity of the peptides.
PSMα3 amyloid-catalyzed hydrolysis was also recorded for several clinically relevant β-lactam antibiotics, and in physiological bacterial settings, underscoring an apparent broad scope of the phenomenon. We observed, for example, PSMα3-mediated catalysis of amoxycillin, a β-lactam antibiotic used for treatment of severe infections14 and is found on the list of essential medicines of the World Health Organization15, and penicillin-G, one of the most widely used antibiotics14. Overall, the observation that functional bacterial amyloids catalyze degradation of common antibiotics may unveil a mechanism for antibiotic bacterial resistance, exposing a new role of functional amyloids in bacterial biofilms. Indeed, different than known β-lactamases that are generally substrate-specific, bacterial amyloids are more promiscuous, making amyloid catalysis pertinent for a wide range of β-lactam antibiotics. An intriguing possibility is that functional bacterial amyloids may have evolved prior to β-lactamases, acting as primitive enzymes in the prebiotic world.
References:
- Hernando-Amado, S., Coque, T. M., Baquero, F. & Martínez, J. L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442 (2019).
- Lima, L. M., Silva, B. N. M. da, Barbosa, G. & Barreiro, E. J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 208, 112829 (2020).
- Otzen, D. & Riek, R. Functional amyloids. Cold Spring Harb. Perspect. Biol. 11, (2019).
- Landau, M. Mimicking cross-α amyloids. Nat. Chem. Biol. 14, 833–834 (2018).
- Tayeb-Fligelman, E. et al. The cytotoxic Staphylococcus aureus PSMα3 reveals a cross-α amyloid-like fibril. Science (80-. ). 355, 831–833 (2017).
- Arad, E. & Jelinek, R. Catalytic amyloids. Trends Chem. (2022) doi:10.1016/j.trechm.2022.07.001.
- Arad, E., Baruch Leshem, A., Rapaport, H. & Jelinek, R. β-Amyloid fibrils catalyze neurotransmitter degradation. Chem Catal. 1, 908–922 (2021).
- Arad, E., Yosefi, G., Bitton, R., Rapaport, H. & Jelinek, R. Native glucagon amyloids catalyze key metabolic reactions. (2022).
- Cheung, G. Y. C. et al. Insight into structure-function relationship in phenol-soluble modulins using an alanine screen of the phenol-soluble modulin (PSM) α3 peptide. FASEB J. 28, 153–161 (2014).
- Joo, H.-S. S., Cheung, G. Y. C., Otto, M., C Cheung, G. Y. & Otto, M. Antimicrobial Activity of Community-associated Methicillin-resistant Staphylococcus aureus Is Caused by Phenol-soluble Modulin Derivatives. J. Biol. Chem. 286, 8933–8940 (2011).
- Viswanatha, T., Marrone, L., Goodfellow, V. & Dmitrienko, G. I. Assays for Β-Lactamase Activity and Inhibition. in Methods in molecular medicine vol. 142 239–260 (2008).
- Morten, M. J. et al. Quantitative super-resolution imaging of pathological aggregates reveals distinct toxicity profiles in different synucleinopathies. Proc. Natl. Acad. Sci. 119, 1–12 (2022).
- de Waal, G. M. et al. Correlative Light-Electron Microscopy detects lipopolysaccharide and its association with fibrin fibres in Parkinson’s Disease, Alzheimer’s Disease and Type 2 Diabetes Mellitus. Sci. Rep. 8, 1–12 (2018).
- Mora-Ochomogo, M. & Lohans, C. T. β-Lactam antibiotic targets and resistance mechanisms: from covalent inhibitors to substrates. RSC Med. Chem. 12, 1623–1639 (2021).
- World Health Organization (WHO). World health organization model list of essential medicines. Ment. Holist. Heal. Some Int. Perspect. 21, 119–134 (2019).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in