Starving Glutamine Selectively in Cancer Cells: Artificial Intelligence Predicted Structure-Based Drug Discovery of Cancer Metabolism Targeting Small Molecule
Published in Biomedical Research
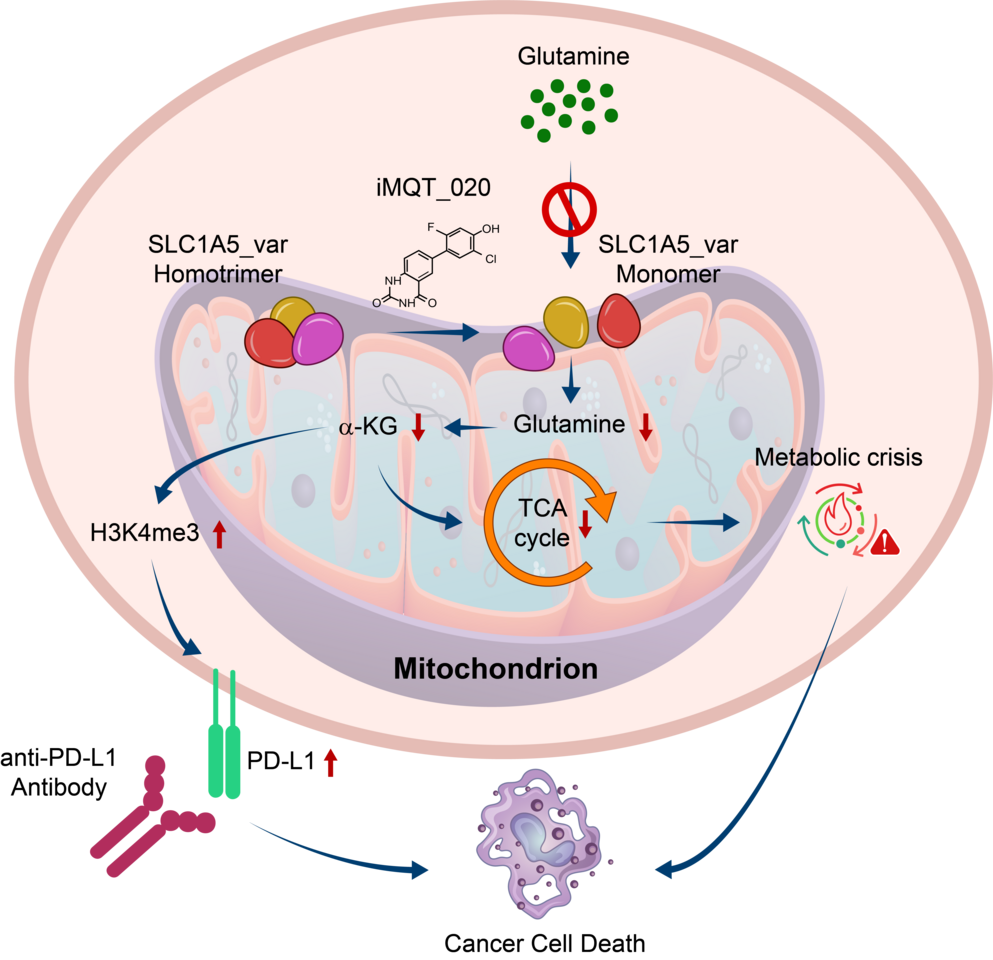
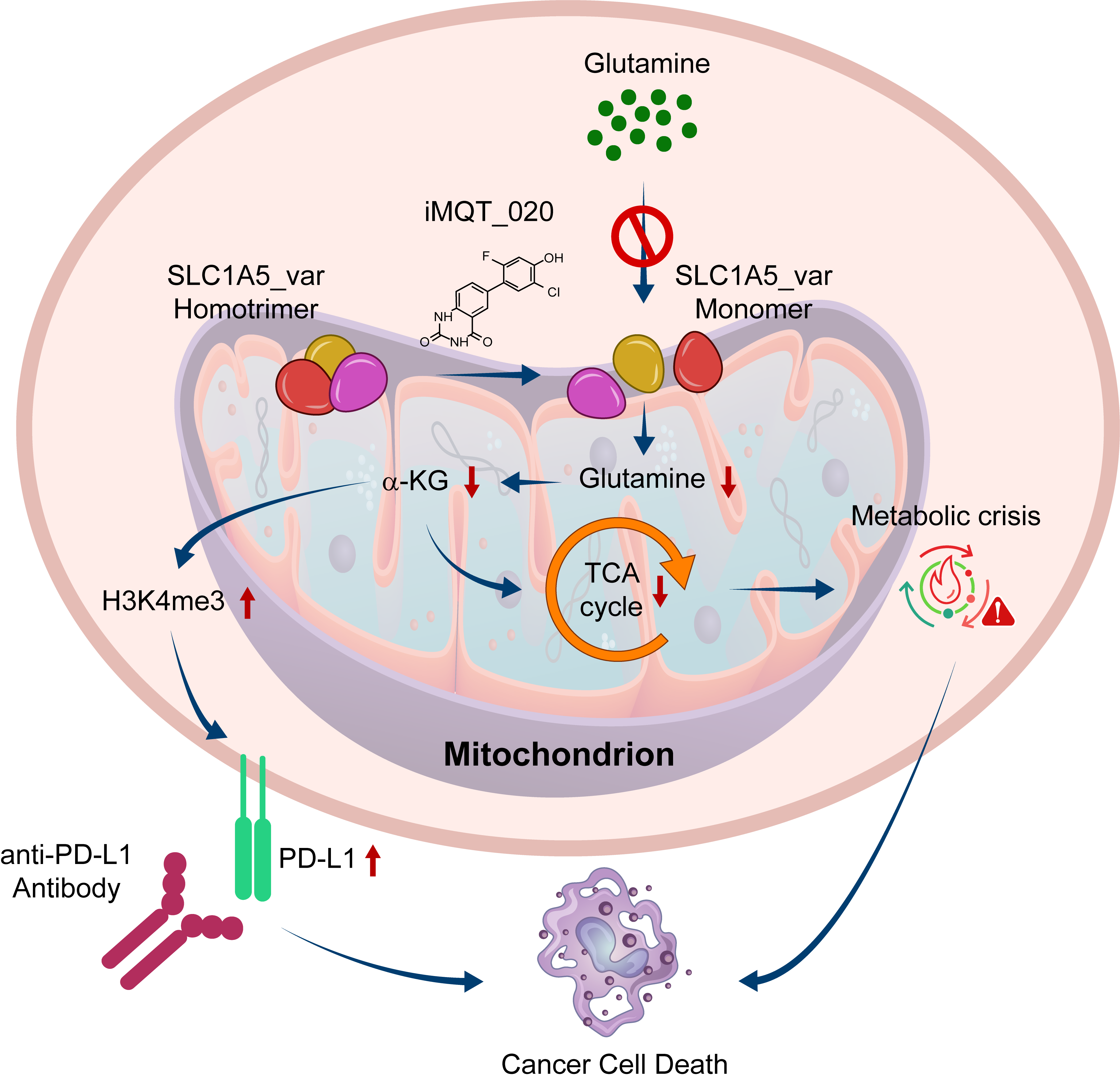
For many years, cancer metabolism has been a fascinating yet daunting frontier. Unlike normal cells, cancer cells often rewire their metabolism to fuel uncontrolled growth. Among these metabolic addictions, glutamine stood out as a prime candidate. Tumor cells consume glutamine at extraordinary rates, making it a lifeline for their survival. But designing drugs that selectively block this dependency without harming healthy cells remained an unsolved challenge. In 2020, our team discovered a variant of SLC1A5 that is localized in the mitochondria and is the major mitochondrial glutamine transporter which takes a critical role in cancer metabolic reprogramming. As this protein was expressed highly only in cancer cells, we began this project, to selectively inhibit this protein.
Harnessing AI to Reimagine Drug Discovery
For correct drug discovery, we initiated a structure-based drug discovery platform. As SLC1A5, the major transcript of the mitochondrial form, was the first protein to have all of its conformation elucidated through Cryo-EM, we thought that validating the structure of SLC1A5 variant would be possible. However, it was hard to gain enough concentration of the protein for structural determination, which led to a completely new project. So, we asked ourselves, are there any other ways to conduct structure-based drug discovery without the structure? We asked, could structure prediction be accurate?
At the time, SwissModel and Homology Modeling were started to solidify their research so we tried to use them too. To check if the model is correct, we synthesized numerous mutants, over 100 different clones with different mutations, so that we could trust the model and conduct structure-based drug screening. Unfortunately, all models came out to be wrong which took quite awhile in this project.
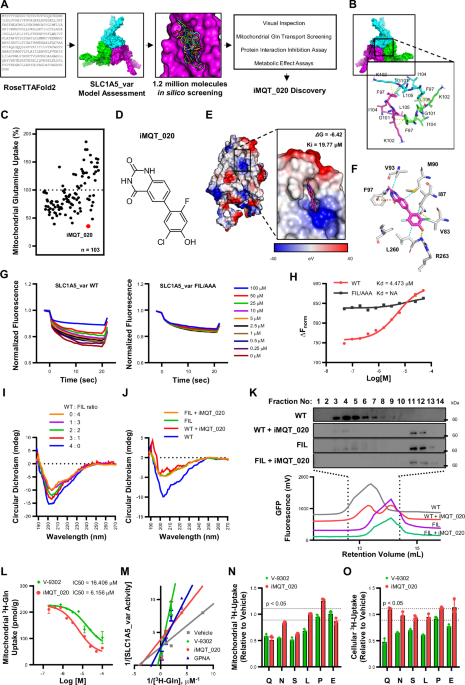
During the journey to find different platforms of de novo and homology protein structure modeling, we came upon the report of Google Deepmind team about AlphaFold. Subsequently we came across RoseTTAFold of Dr. Baker, a future nobel prize winner, and we started learning the principles of artificial intelligence. And we asked could artificial intelligence (AI) accelerate this process? In brief, among 30 models predicted by the AI, 1 came out to be correct according to our biochemical experiments.
A Drug That Starves Tumors, But Spares Healthy Cells
With the correct model structure via AI, we sped things up. By integrating deep neural networks, protein structure prediction, and virtual drug screening, we rapidly analyzed over 1.2 million compounds. The target: the mitochondrial glutamine transporter, a gatekeeper that fuels cancer cell metabolism. Within months, not years, our AI models highlighted a handful of promising molecules predicted to bind with high affinity. What followed was a careful cycle of validation — computational predictions guiding wet-lab assays, and experimental results refining our models. Slowly but surely, the contours of a viable drug candidate began to emerge.
Fortunately, our best compound showed a remarkable property: it selectively suppressed the growth of pancreatic and other hard-to-treat solid tumor cells, while leaving normal cells largely unaffected. In preclinical models, the compound effectively slowed tumor progression. Even more strikingly, we uncovered a new dimension of its activity: inhibiting glutamine metabolism reshaped the tumor microenvironment in ways that boosted the effectiveness of PD-L1 immune checkpoint blockade. For patients who previously failed to respond to immunotherapy, this combination strategy could open new therapeutic doors.
Lessons Learned and the Road Ahead
This journey taught us that AI is not just a buzzword in drug discovery — it can genuinely transform the way we search for first-in-class therapies, cutting down both time and cost while expanding our imagination of what’s possible. Of course, this is only the beginning. Much work lies ahead to refine our compound, understand its mechanisms in greater depth, and ultimately translate it into a clinically approved therapy. But we believe this study — now published in Nature Communications — represents a step toward a new paradigm: precision metabolic therapies powered by AI.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Thank you for sharing!