Statistically unbiased prediction enables accurate denoising of voltage imaging data
Published in Bioengineering & Biotechnology

Voltage imaging is a functional imaging technique used to optically measure the electrical activity of neurons. It employs certain fluorescent molecules whose fluorescence intensity changes in response to the electrical potential, allowing the fluorescence signals from the membrane to directly encode the transmembrane potential information. Our team has been pushing the boundaries of voltage imaging towards volumetric and large-scale imaging, which would enable us to monitor how a large number of neurons in an intact brain communicate with other neurons with high spatiotemporal precision. Unfortunately, our attempts only highlighted the many technical challenges associated with it. The primary issue in scaling voltage imaging was the signal-to-noise ratio (SNR). Given that our signal of interest — the action potential — changes on a millisecond-scale, the imaging must be conducted at or near kHz frequencies, which restricts the number of available photons per frame to two orders of magnitude lower than that in calcium imaging. Moreover, the amplitude of the signal (dF/F) in voltage imaging is about an order of magnitude lower than that in calcium imaging. Overall, these factors posed a significant challenge in terms of SNR for advancing voltage imaging.
Considering the recent success achieved by DeepCAD and DeepInterpolation, which increased the SNR of calcium imaging data tenfold, it was natural for us to attempt to apply them to address the SNR issue. Unfortunately, we were unable to detect the spikes associated with the action potentials post-denoising. Initially, we were uncertain whether this failure was due to a lack of neuronal activity in the animal or another factor. Following multiple trials, it became clear that the spikes did not “survive” the denoising process, prompting us to investigate further. After some reflection, we realized that this outcome was unavoidable. These methods rely on predicting the current frame using information from temporally adjacent frames, whereby the predictable component, which is presumably the signal, “passes” through the network, while the unpredictable component, presumed to be noise, is attenuated by the network. Consequently, unless the data were oversampled, the spikes were attenuated, as they were not predictable based on the information from temporally adjacent frames.
While seeking a solution, we realized that the relevant information for denoising a pixel in a given frame resides within its spatial neighbors in the same frame which was discarded during the DeepCAD and DeepInterpolation processes. Theoretically, we could discard a single pixel in one frame of the video and predict its value using the remainder of the video, thereby accurately discerning the true signal. We devised an optimal convolutional network architecture that performs such operations in parallel, introducing the concept of a spatiotemporal blind spot. After experimenting with several variants, eventually everything functioned seamlessly. The algorithm could denoise all the voltage imaging data we encountered while retaining the sharp transient signals. In light of its capacity to leverage spatiotemporal dependencies among pixel values for denoising, and anticipating its potential to support the research community, we have named it SUPPORT (Statistically Unbiased Prediction utilizing sPatiOtempoRal information in imaging daTa).
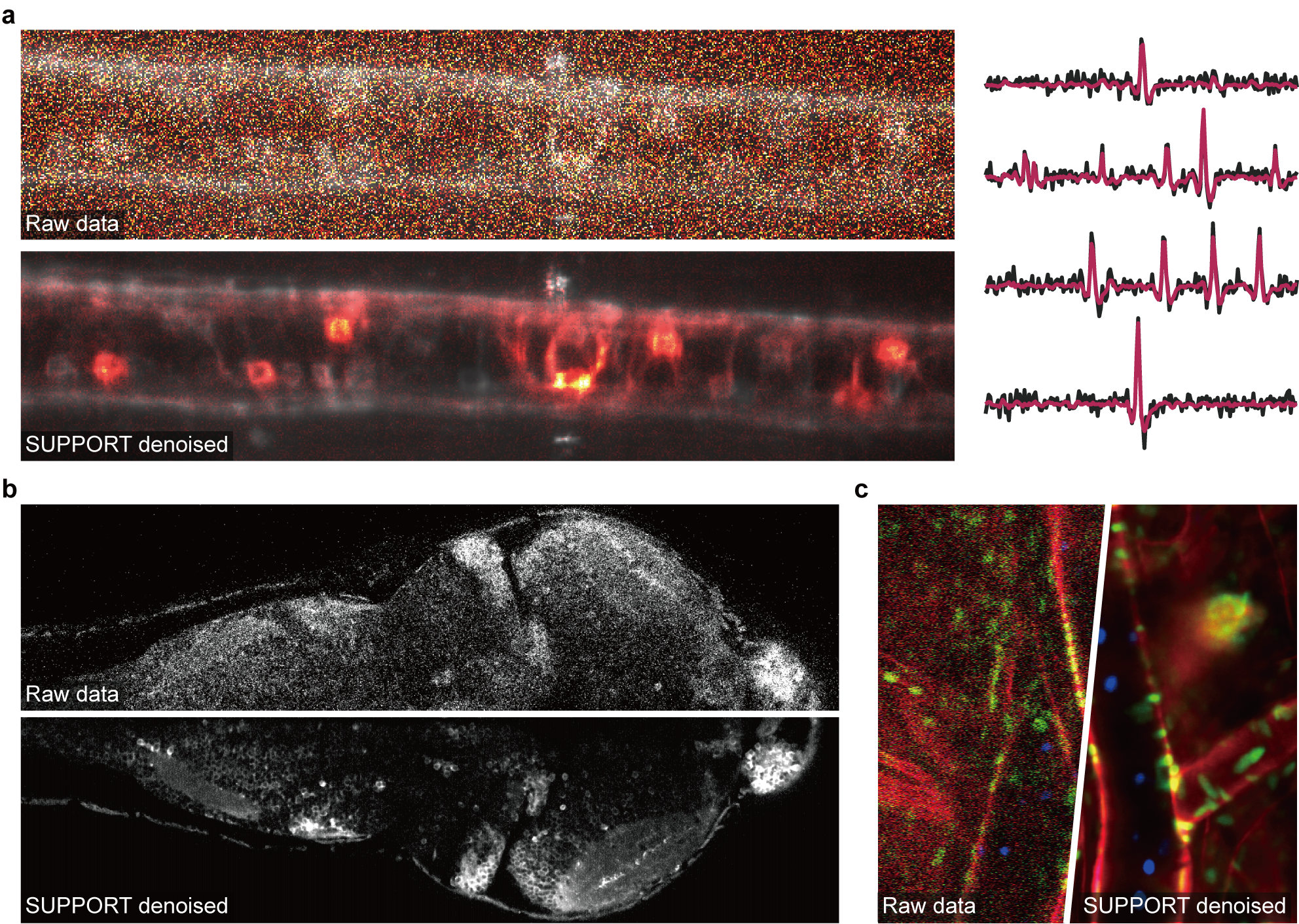
Figure 1. Denoising various images using SUPPORT. (a) Voltage imaging of a zebrafish spinal cord expressing zArchon1. Top-left: Raw data. Bottom-left: SUPPORT-denoised data. Right: Extracted traces from raw (black) and SUPPORT-denoised (red) data. (b) Calcium imaging of a zebrafish whole brain expressing GCaMP7a. (c) Intravital imaging of mouse ear skin.
We were especially fascinated by the results we obtained by applying SUPPORT to voltage imaging data from larval zebrafish, in which both the image itself and the extracted signals were greatly improved (Fig. 1a). We then realized that SUPPORT is ideal not only for voltage imaging but also for calcium imaging, because not all neuroscientists have access to or wish to use an imaging system that is fast enough to oversample the calcium dynamics. As expected, SUPPORT was perfectly capable of denoising calcium imaging data that was acquired at only a few hertz (Fig. 1b).
Later on, we discovered that for a specific dataset from our collaborator, our network produced output that was nearly identical to the input, indicating an inability to denoise the data. This suggested that just by knowing the spatiotemporal neighboring pixels of a given pixel, the network can accurately predict the value of the given pixel, including noise. This meant the noise in each pixel was not independent of its spatiotemporal neighbors. Initially, we were puzzled because this occurred only with datasets from a specific microscope and not with others. We identified that the images from the microscope used to acquire the data contained certain horizontal patterns, as wide as 10 pixels, which were suspected to be the cause. We speculated that this might be due to the combination of fast horizontal scanning and the low-frequency noise in the detector, such as 1/f noise, which causes the noise in each pixel to depend on its spatial neighbors. We reasoned that since such noise correlation is limited to a small local area, the network would not be able to predict the noise in a pixel if we obscured a 1×19 pixel area around it. Fortunately, our network architecture was exceptionally suited for programming the size of the blind spot, even though this was not initially intended in its design. The results confirmed our speculation (Fig. 1c).
We believe our work will find immediate applications in the broader neuroscience and bio-imaging communities. This is because, unlike most other recent advances in machine learning applications for imaging and image analysis, our method does not necessitate any specific experimental setting or training data. Our technique can significantly enhance any type of functional imaging data, time-lapse imaging data, or even static volumetric images, regardless of whether it was taken with sufficient imaging speed or not.
Lastly, we would like to express our team’s sincere appreciation to the editor and the reviewers for their constructive and critical feedback. They recommended, for example, that we concentrate on the voltage imaging application, conduct extended validation on a large number of datasets, and examine how SUPPORT performs on recordings containing motion. We believe that these suggestions were pivotal in verifying SUPPORT’s capabilities and in strengthening the manuscript.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in