Tackling trypanosome biology takes a lot of guts
Published in Microbiology

There’s a special kind of friendship that comes from sitting side-by-side plucking the heads off sleepy tsetse flies. In a three year long collaboration between the Hill laboratory at the University of California, Los Angeles and the Roditi laboratory at the University of Bern, Sebastian Shaw and I dissected a lot of tsetse flies.
Both the Hill and Roditi labs study the protozoan parasite, Trypanosoma brucei (T. brucei), which infects mammals, causing African sleeping sickness in humans and Nagana in cattle, and is transmitted by tsetse flies. In the Hill lab we are interested in understanding how T. brucei senses and transduces signals. We had previously found that when T. brucei lost a particular cAMP-specific phosphodiesterase, they could no longer engage in a group behavior called social motility, which is used as readout for signaling capability of the parasite in vitro. With the Roditi lab’s expertise in T. brucei and tsetse fly biology, we decided to work together to ask the question, what is the role of cAMP signaling in T. brucei during tsetse fly infection?
On three separate trips to Bern, Sebastian and I infected tsetse flies with wild type or phosphodiesterase-B1 knockout (PDEB1 KO) parasites. At first, we were only interested in asking if the mutant parasites were able to infect the midgut of the tsetse fly, the first site of parasite infection in the fly, but Sebastian suggested that we should also take a look at the proventriculus, the next organ that parasites infect on their journey to the salivary glands.
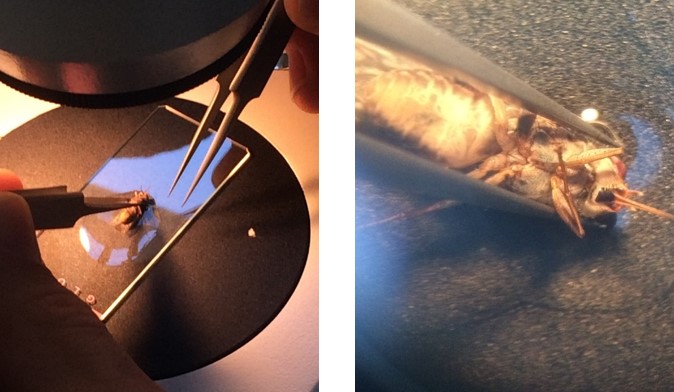
Figure 1: Tsetse fly dissection. (Left) Under the microscope. (Right) Up close!
We dissected flies and assessed their level of infection. If a fly midgut was heavily infected with parasites, we also checked the infection level of the proventriculus. Pretty soon we fell into a rhythm. Pick up a sleeping fly, rip off its head, open it up, pull out the midgut and proventriculus, and check for parasites under the microscope. As we dissected flies infected with PDEB1 KO parasites, Sebastian, who was on the microscope, was taking longer than usual.
Finally, he said, “I don’t think it’s infected. I can’t find any parasites.”
This was huge. Of the flies infected with PDEB1 KO parasites, all of the flies, except for one, had no parasites present in their proventriculus. In the one fly that did, we saw only one lonely parasite swimming around. Clearly, cAMP signaling is necessary for T. brucei to move from the fly midgut to the proventriculus.
But where exactly is this defect between the midgut and the proventriculus? To move from the midgut to the proventriculus, T. brucei must get to the other side of the peritrophic matrix, a chitinous structure separating the lumen of the midgut from the ectoperitrophic space. It is in this ectoperitrophic space that T. brucei migrate to the proventriculus.
By feeding flies fluorescent parasites and a fluorescent stain that would coat the peritrophic matrix, dissecting intact midguts, and adding a DNA stain, we could tell if parasites were stuck inside the midgut or had moved into the ectoperitrophic space.
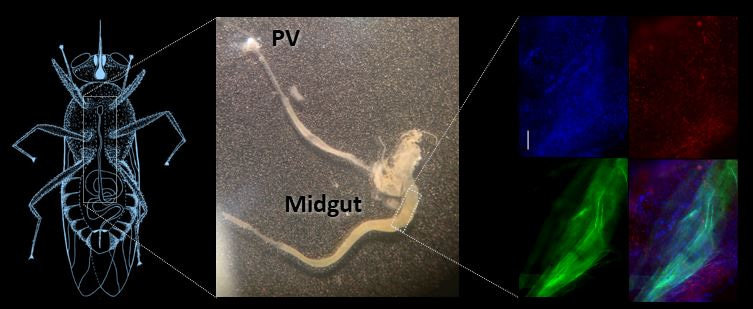
Figure 2: Parasites within. (Left) Drawing of a tsetse fly. (Middle) Midgut and proventriculus segment of the fly’s GI tract. (Right) Trypanosomes (red) in the tsetse fly midgut (blue) with the peritrophic matrix labeled in green. (Left and Right images adapted from: https://rdcu.be/bniGa )
This procedure was one of the toughest. Normal midgut and proventriculus fly dissections might take a couple of minutes per fly. These whole midgut dissections, however, could take up to 10-15 minutes per fly. If we made a single rip or tear in the midgut, we could not be sure whether the parasites had spilled into the ectoperitrophic space from the rip or if they were there all along.
An extra stress on an already difficult procedure came in the way of an unseasonably cold winter in Switzerland. Just as our pupae of flies were arriving from Bratislava and I was arriving from California, so did temperatures of –10°C, killing most of our flies. Luckily, our fly supplier was kind enough to quickly ship us their extra fly pupae, and our experiment went on as planned.
From the confocal images of our careful dissections, we saw that PDEB1 KO parasites have a defect in moving out of the midgut lumen and into the ectoperitrophic space, indicating that cAMP signaling is necessary for this step in the T. brucei transmission cycle.
These exciting results would not have been possible without the teamwork and friendship of Sebastian and myself, our advisors, and our lab mates. Whether it was suggesting a coffee or pizza break during late night microscopy sessions, we kept each other going and pushed the T. brucei field forward in the process.
The results of our work can be found in our recently published paper in Nature Communications: Flagellar cAMP signaling controls trypanosome progression through host tissues. https://rdcu.be/bniGa
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in