Targeting large DNA sequences in human pluripotent stem cells
Published in Bioengineering & Biotechnology, Microbiology, and Protocols & Methods

Human pluripotent stem cells (hPSCs) hold unparalleled potential in medicine due to their ability to differentiate into any cell type in the human body. This means when hPSCs carry specific genetic mutations, the differentiated cells also carry the same mutations, making them ideal for studying disease mechanisms and investigating potential therapeutic applications. Since 2012, CRISPR/Cas9 technology1 has revolutionized these studies by enabling precise corrections of mutations in patient-derived hiPSCs or introducing specific mutations to model diseases. While effective, the process remains time-consuming, often requiring six months and involving tedious procedures such as screening numerous clones to identify suitable cell lines for studying the disease.
Another challenge with CRISPR/Cas9 is that it often lacks precise control over which allele is modified. This issue is particularly problematic in diseases where additional polymorphisms within the same gene can have distinct impacts on the disease phenotype. For example, our research into cardiac arrhythmias demonstrated how the allele carrying a common polymorphism, in relation to the primary genetic mutation, significantly influenced the severity and treatment responsiveness2. This highlighted the need for technologies that enable precise allele-specific modifications, whether for introducing gene variants or reporter genes to study gene function.
In 2018, we along with Catarina Grandela, a Postdoc in the group, set out to develop a solution. Our goal was to streamline the integration of large DNA segments into hPSCs while ensuring precision and efficiency. The outcome was STRAIGHT-IN (Serine and Tyrosine Recombinase Assisted Integration of Genes for High-Throughput Investigation), a platform that combines CRISPR/Cas9 technology with recombinase systems to enhance our ability to generate targeted genetically modified hiPSCs 3.
STRAIGHT-IN employs three key enzymes from the synthetic biology toolbox:
(1) CRISPR/Cas9, which targets and prepares specific genomic sites for integration by inserting a “landing pad”.
(2) Serine recombinases, such as Bxb1, which integrate large DNA payloads into these pre-targeted sites.
(3) Tyrosine recombinases, like Cre, which remove most auxiliary elements needed in the first two steps, minimizing genomic disruption and scarring.
This procedure not only meant that we could reliably perform the targeted replacement of DNA sequences up to 50 kb in length (even achieving a targeting of 170 kb), but it also drastically reduced the time required to develop the genetically modified hiPSC lines—from months to just weeks.
As we were developing the STRAIGHT-IN platform, we realized its potential also for synthetic biology applications. The platform's ability to integrate reporters, biosensors, or complex genetic circuits into hiPSCs (Figure) with high precision and repeatability is essential for the design-build-test-learn cycles typical in synthetic biology. This capability ensures consistent and reliable outcomes, crucial for rapid prototyping and iterative experimentation in synthetic biology.
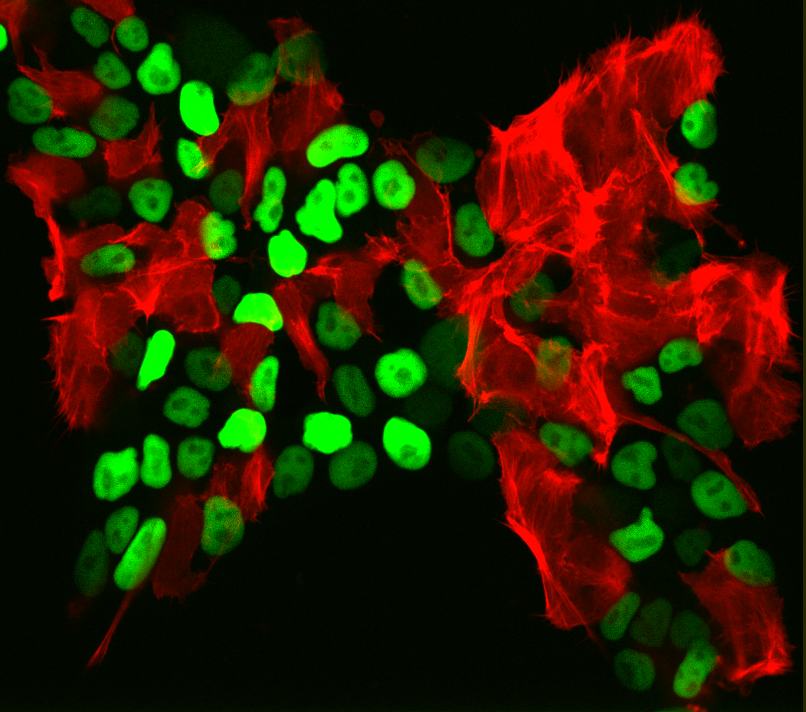
hiPSCs after STRAIGHT-IN procedure in which two fluorescent reporters (green; nuclei, red; F-actin) were delivered simultaneously but only one could integrate per cell.
Over the last couple of years, we have focused on refining the landing pads and donor plasmids to increase the efficiency and shorten the time needed to generate hiPSC lines containing the intended genetic material. In 2023, we published the second version of the STRAIGHT-IN platform which allows for 100% enrichment of hiPSCs containing the desired DNA payloads4. These improvements significantly reduce the number of hiPSC clones that need screening, and even enable us to work with bulk populations that are homogenous both genotypically and phenotypically.
With the recent publication of our detailed protocol for using STRAIGHT-IN in Nature Protocols5, we aim to democratize this technology and make it accessible to labs worldwide. To this end, we also have made all necessary plasmids available through Addgene (https://www.addgene.org/Richard_Davis). Researchers can implement our platform in their work without needing specialized equipment or expertise beyond core molecular biology and stem cell culture skills. The protocol is already being adopted by graduate students and postdoctoral fellows in various research groups, both within our institute6 and externally.
The journey from concept to protocol has been incredibly rewarding, and we are continuing to further develop STRAIGHT-IN and expand its applications. We are also excited to see how other researchers utilize this technology to advance their own studies. We believe that STRAIGHT-IN will not only improve scalability in disease modeling, but also accelerate the development of innovative treatments and therapies.
References
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J.A., and Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821.
- van den Brink, L., Brandão, K.O., Yiangou, L., Blanch-Asensio, A., Mol, M.P.H., Mummery, C.L., Verkerk, A.O., and Davis, R.P. (2021). The Linkage Phase of the Polymorphism KCNH2-K897T Influences the Electrophysiological Phenotype in hiPSC Models of LQT2. Front Physiol 12. https://doi.org/10.3389/fphys.2021.755642.
- Blanch-Asensio, A., Grandela, C., Brandão, K.O., de Korte, T., Mei, H., Ariyurek, Y., Yiangou, L., Mol, M.P.H., van Meer, B.J., Kloet, S.L., et al. (2022). STRAIGHT-IN enables high-throughput targeting of large DNA payloads in human pluripotent stem cells. Cell Reports Methods 2, 100300. https://doi.org/10.1016/J.CRMETH.2022.100300.
- Blanch-Asensio, A., Vaart, B. Van Der, Vinagre, M., Groen, E., Arendzen, C., Freund, C., Geijsen, N., Mummery, C.L., and Davis, R.P. (2023). Generation of AAVS1 and CLYBL STRAIGHT-IN v2 acceptor human iPSC lines for integrating DNA payloads. Stem Cell Res 66, 102991. https://doi.org/10.1016/j.scr.2022.102991.
- Blanch-Asensio, A., Grandela, C., Mummery, C.L., and Davis, R.P. (2024). STRAIGHT-IN: a platform for rapidly generating panels of genetically modified human pluripotent stem cell lines. Nat Protoc. https://doi.org/10.1038/s41596-024-01039-2.
- Meraviglia, V., Blanch-Asensio, A., Davis, R., Mummery, C.L., and Bellin, M. (2024). Generation of isogenic allelic series of LMNA-mutated hiPSC lines using the novel and highly-efficient targeting platform, STRAIGHT-IN. Cardiovasc Res 120, i266.
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in