Temporal dynamics and persistence of resistance genes to broad spectrum antibiotics in an urban community
Published in Microbiology, Biomedical Research, and Public Health
The discovery of penicillin by Sir Alexander Fleming in the 1940s was hailed as a medical marvel. However, shortly after receiving the Nobel Prize in an interview, Fleming cautioned, the thoughtless person playing with penicillin treatment is morally responsible for the death of the man who succumbs to infection with the penicillin-resistant organism1. Tragically, this grim forecast, made over seven decades ago, has become our reality as antibiotics lose their efficacy and resistant strains proliferate.
Why AMR Is a Dire Threat to Our Future
Antimicrobial Resistance (AMR), once known as the "silent pandemic," has now become impossible to ignore. It is on the verge of becoming the next major global health crisis, and its effects are becoming increasingly apparent2. It threatens to roll back decades of medical progress, undermine food production systems, and significantly effecting life expectancy. The antibiotics that were once thought to be wonder drugs, used liberally to treat common infections to serious diseases, are now encountering resistance3, it is almost slowly losing their magic touch. If the post-antibiotic era is not addressed, it could claim more lives annually by 2050 than cancer4.
AMR: Beyond the Hospital Walls
Due to its extensive clinical impact, AMR has been primarily addressed within healthcare, focusing on minimizing infections and ensuring patient safety. A key aspect of AMR management is patient-based surveillance, which involves identifying and quantifying AMR from clinical samples5. However, microbes and their resistance genes do not respect boundaries – they move freely between humans, animals, and the environment. This is where the One Health approach comes in. It emphasizes collaboration across disciplines to tackle AMR as a unified problem6. While minimizing infections and tracking resistance in healthcare settings remain critical, a complete understanding of AMR requires venturing beyond hospital walls. AMR patterns within the broader population and environment are crucial for accurate risk assessment5. However, our current knowledge in this area remains limited, hindering our ability to effectively control this growing threat.
Sewage Secrets: Unveiling Antibiotic Resistance with Wastewater
In our recent study, we aimed to bridge this gap by examining the temporal dynamics and persistence of resistance genes to broad-spectrum antibiotics within an urban community. We used wastewater-based epidemiology (WBE), a novel approach that provides valuable insights that are otherwise difficult to obtain. WBE analyses wastewater for the presence of antibiotic resistance genes/bacteria (ARGs/ARBs) in the context of community.
Why WBE Matters
This technique goes beyond traditional methods. WBE is ethical, cost-effective, and gathers data from selected population/community, including those with limited healthcare access8,9. It also pinpoints AMR hotspots in a specific location, allowing us to identify areas of concern, track trends, and compare resistance levels across different groups. WBE's power extends beyond early detection. By analysing wastewater, we can help medical professionals choose the most effective antibiotics for their patients. Additionally, WBE data helps us evaluate wastewater treatment efficacy and improve sanitation practices, ultimately helping to reduce the environmental spread of AMR7,8,9.
The Study Approach
To explore the resistance dynamics and prevalence of ARGs (123) and MGEs, (13) we conducted a detailed analysis using quantitative real-time PCR (q-PCR) in the wastewater of a selected urban community. Our sampling was carried out monthly over a five-month period, from December 2021 to April 2022. This approach allowed us to capture the resistance diversity, temporal dynamics, co-abundance of ARGs and MGEs, and the mechanisms underlying these resistances.
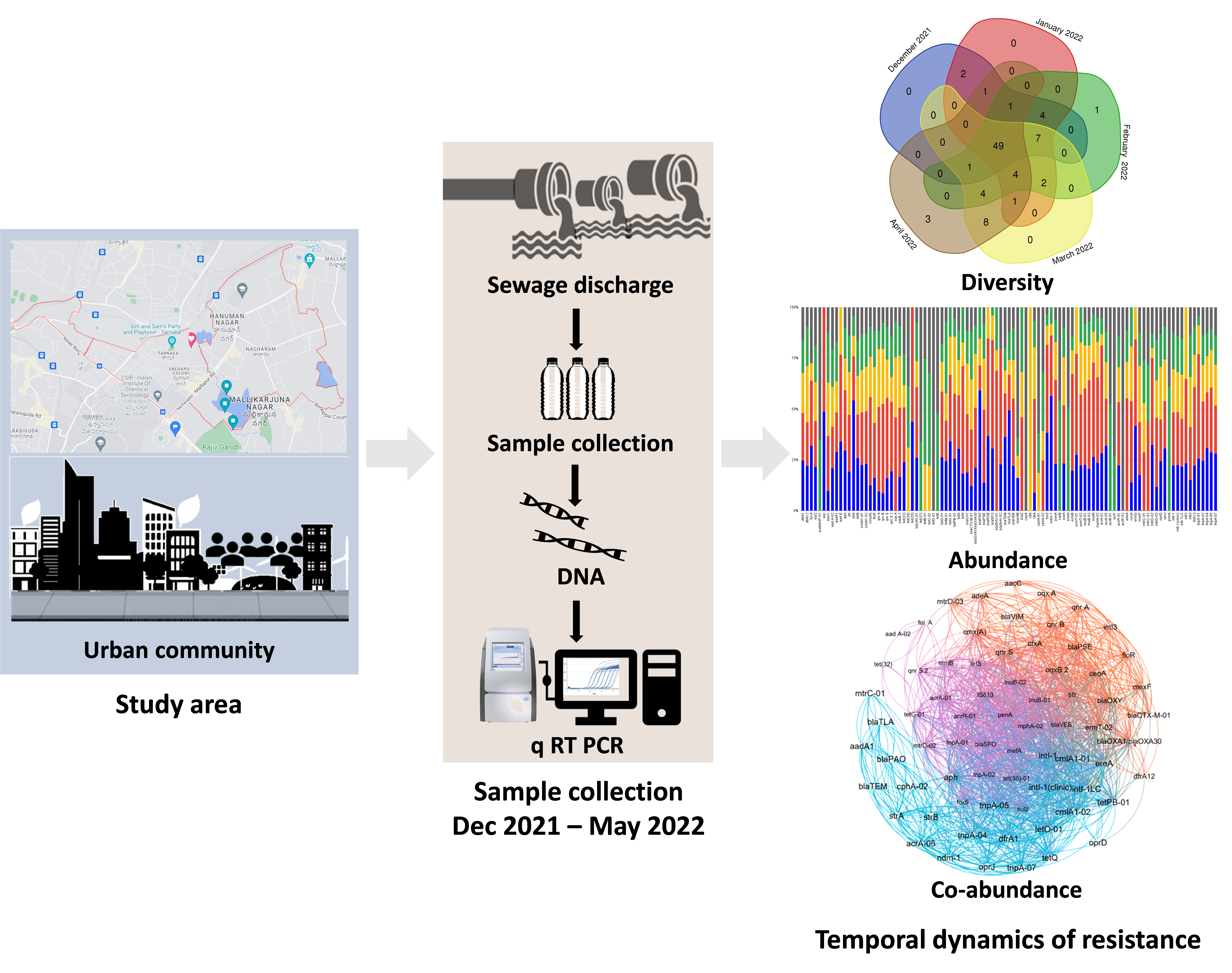
Key Findings
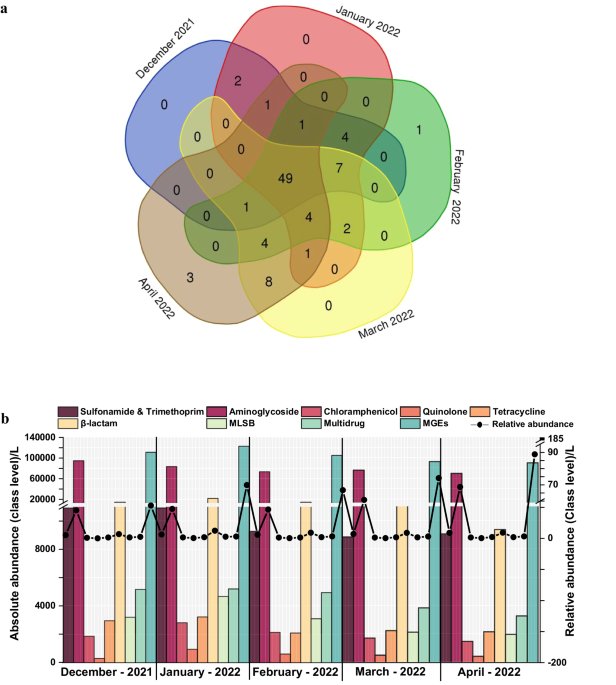
- Consistent Detection of ARGs: One of the most striking findings was that approximately 50% of the tested ARG subtypes were detected consistently across all months. The detection frequencies ranged from 52% to 61%, with the highest absolute abundance observed in the winter months.
- Co-Abundance and Module Clustering: Co-abundance analysis revealed that specific genes clustered into distinct modules, highlighting shared distribution patterns and functional associations among ARGs and MGEs. This modular structure indicates complex interactions and co-evolution of resistance genes within the community.
- Clinically Significant Genes: Detected clinically significant genes such as ndm-1 and cfiA, along with other variants like blaoxy, aph, aacC, tet-35, tet M, and tet-32. These genes are capable of conferring resistance to 3rd and 4th generation β-lactam, aminoglycoside, tetracycline, and multi-drug classes, significantly contributing to core/persistent resistance.
- Seasonal Fluctuations: Study uncovered that the winter months showed the highest absolute abundance of ARGs. This seasonal trend is crucial for understanding the temporal dynamics of resistance gene distribution and could inform targeted interventions during the peak periods.
Conclusion
The temporal dynamics and persistence of resistance genes provides a crucial window into the broader AMR landscape. Our study on urban AMR dynamics, utilizing proxy wastewater analysis, offers crucial insights for controlling the spread of antibiotic resistance. Understanding the persistence and location of resistance genes allows us to target interventions, such as promoting responsible antibiotic use, where they are most effective. This knowledge is essential useful for informing policy and developing strategies to mitigate AMR's impact on public health. For a deeper dive, see our full research in npj Clean Water. Together, let us combat AMR and ensure a healthier future.
Key Takeaways
- Wastewater analysis offers insights for AMR control.
- Continuous monitoring and data collection are essential.
Social Networks and Webpages:
X: @SVenkataMohan2; @JavvadiYamini
Google Scholar: https://scholar.google.com/citations?hl=en&user=gJa0fbsAAAAJ
IICT: https://www.iict.res.in/PERSONAL_PROFILE
References
- Rosenblatt-Farrell N. The landscape of antibiotic resistance. Environ Health Perspect. 117, A244-50 (2009).
- Gautam, A. Antimicrobial resistance: the next probable pandemic. JNMA, 60, 225 (2022).
- Hay et al. Measuring and mapping the global burden of antimicrobial resistance. BMC Med, 16, 1-3 (2018).
- Murray et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet, 399, 629-655 (2022).
- Hart et al. Environmental surveillance of antimicrobial resistance (AMR), perspectives from a national environmental regulator in 2023. Euro Surveill, 28, 2200367 (2023).
- Collignon et al. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis Lancet Planet Health 2, e398–e405 (2018).
- Javvadi, Y. and Mohan, S.V. Understanding the distribution of antibiotic resistance genes in an urban community using wastewater-based epidemiological approach. Total Environ. 868, 161419 (2023)
- Hendriksen et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 10, 1–12 (2019).
Mohan et al. SARS-CoV-2 in environmental perspective: occurrence, persistence, surveillance, inactivation, and challenges. Chem. Eng. J. 405, 126893 (2021).
Follow the Topic
-
npj Clean Water

This journal publishes high-quality papers which report cutting-edge science, technology, application, policy and societal issues that contribute towards a more sustainable supply of clean water.
Related Collections
With Collections, you can get published faster and increase your visibility.
Advancing Artificial Intelligence Innovations for Resilient and Sustainable Clean Water: Novel Methodologies and Case Studies
Publishing Model: Open Access
Deadline: Feb 28, 2026
Advances in Smart Water Systems: From Source to Tap and Beyond
Publishing Model: Open Access
Deadline: Feb 28, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in