The birth of the T7-DIVA system for continuous evolution of proteins
Published in Bioengineering & Biotechnology
In our laboratory E. coli is the workhorse for projects like the selection of small antibody fragments [1] or the expression of complex nanomachines for protein secretion [2, 3]. We commonly face issues like antibodies having low affinity or protein complexes with suboptimal assembly, which often make us think of improving the funciontal properties of these proteins. But common in vitro mutagenesis techniques are tedious and time-consuming, requiring the cloning of large libraries of the mutagenized gene(s) in iterative cycles.
Back in 2015 we started to look for an in vivo mutagenesis system that could be easily implemented for the continuous evolution of genes with different lengths, from short gene segments to large operons. We found none so versatile and started to think of its design. We knew of the mutagenic activity of various cytosine deaminases (CDs) when expressed in E. coli [4] and started to entertain the idea of tethering CDs to the catalytically dead Cas9 (dCas9) that can be directed with CRISPR RNA (crRNA) or gRNA. But soon the works of Komor et al. [5] and Nishida et al. [6] demonstrated that this strategy induces mutations in a narrow window of bases within the binding region of dCas9. It was not the way to go for us. Instead, we thought of linking the CDs to a processive enzyme like the T7 RNA polymerase, so that the mutagenic enzyme could move along the gene. As we wanted to preserve endogenous expression of the target gene, we placed the T7 promoter downstream of the gene in reverse orientation (Figure 1). We started our work by fusing the human CD AID [7] to T7RNAP and constructed an E. coli strain carrying the yeast URA3 gene in the chromosome as a way to quantify the mutagenic activity of the fusion. When we observed that AID fused to T7RNAP induced mutations in URA3 having the T7 promoter, we explored other CDs, like rAPOBEC1 from rat and pmCDA1 from lamprey [4], and a recently reported adenosine deaminase (AD) [8], as partners for fusion to T7RNAP. They all worked!
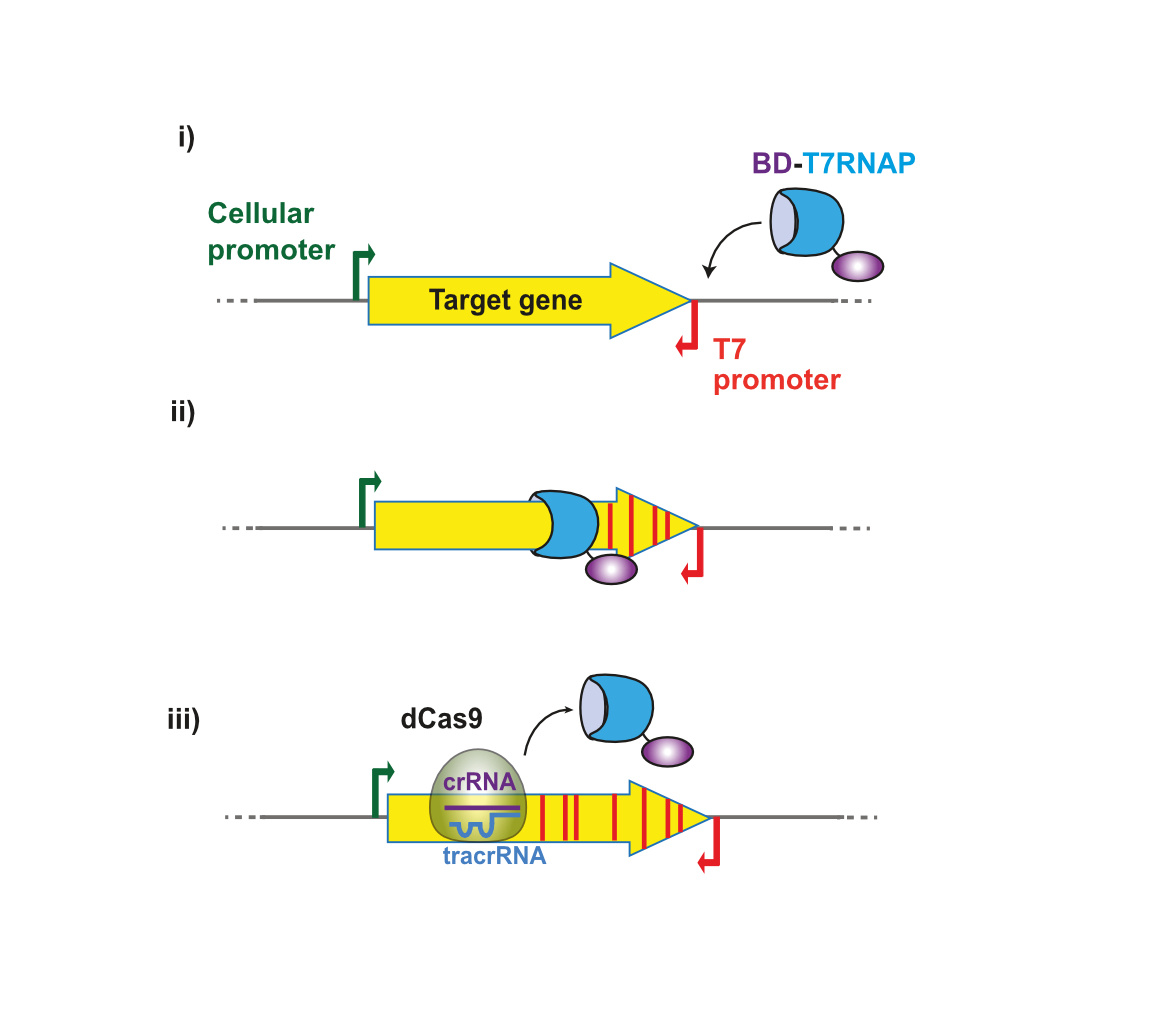 Figure 1. Schematic representation of the mutagenesis process by T7-DIVA
Figure 1. Schematic representation of the mutagenesis process by T7-DIVA
Then an independent group reported that a chimeric fusion between rAPOBEC1 and T7RNAP, named Muta-T7, was able to mutagenize in E. coli an antibiotic resistance gene cloned in a plasmid [9]. They described the high processivity of Muta-T7 fusion beyond the target gene, so at least four T7 transcriptional terminators were needed to prevent mutagenesis of downstream genes. But this approach cannot be used to mutagenize a gene segment because the arrays of terminators will truncate the gene. We proposed a much more flexible solution to this problem. We knew dCas9 was able to block elongation of the bacterial RNA polymerase [10] but... was it sufficient to stop the potent T7RNAP and consequently delimit the mutagenesis window between the T7 promoter and the binding site of dCas9? Luckily for us it also worked, albeit it was necessary three dCas9 molecules in a row to stop the AID-T7RNAP fusion.
We named our system T7-DIVA (T7-targeted dCas9-limited in vivo mutagenesis) and tested it for the continuous evolution of the b-lactam resistance gene TEM-1. Only two cycles of growth and induction of the mutagenic enzyme and dCas9 for protecion of downstream genes were sufficient to evolve TEM-1 mutants with resistance to high concentrations of a third-generation cephalosporine (Figure 2). We think T7-DIVA can be readily applied in multiple molecular evolution processes of proteins of interest, like enzymes and antibodies, and for the engineering of complex metabolic pathways found in operons (Figure 3).
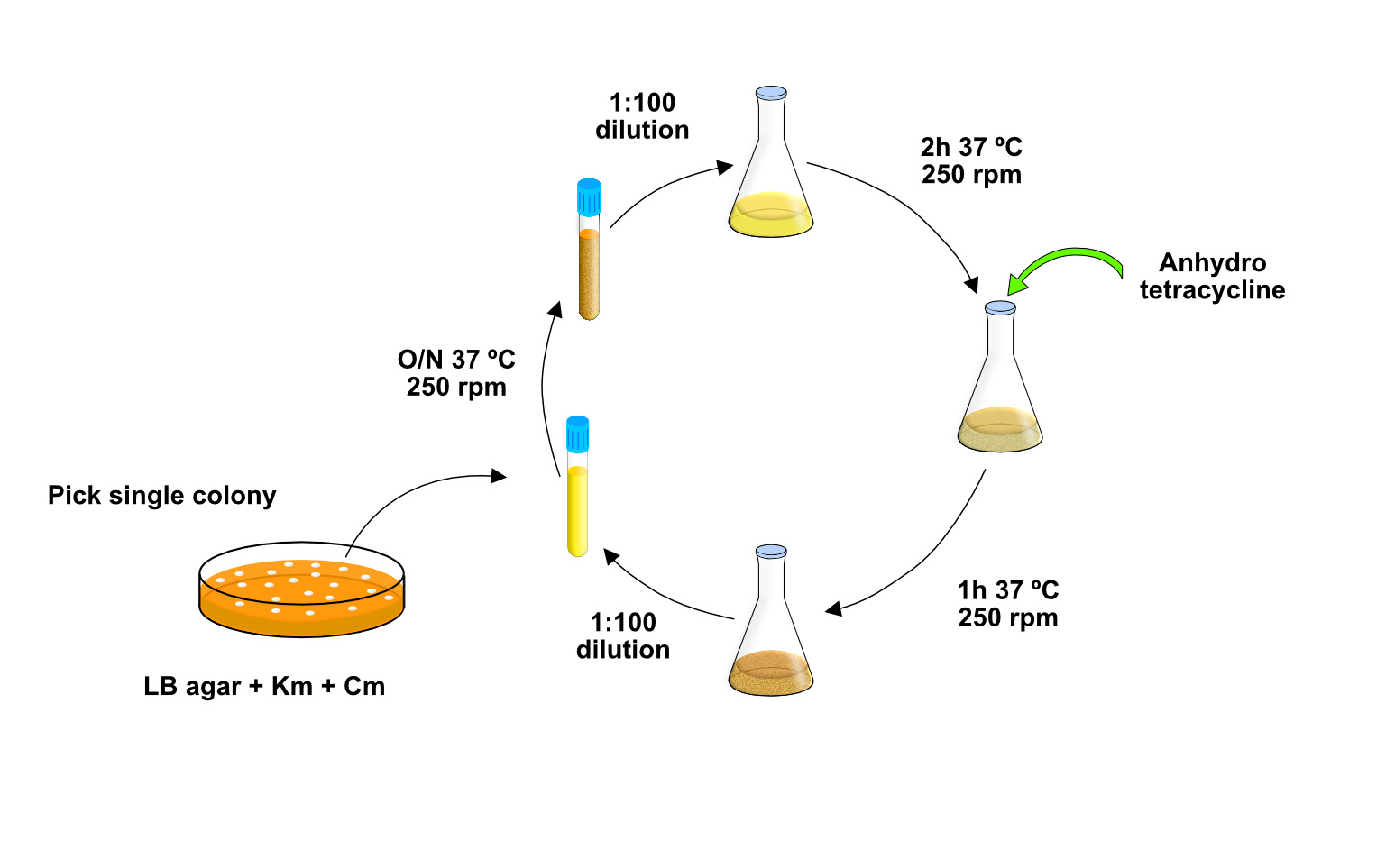 Figure 2. Easy and fast experimental design for the continuous evolution process with T7-DIVA system
Figure 2. Easy and fast experimental design for the continuous evolution process with T7-DIVA system
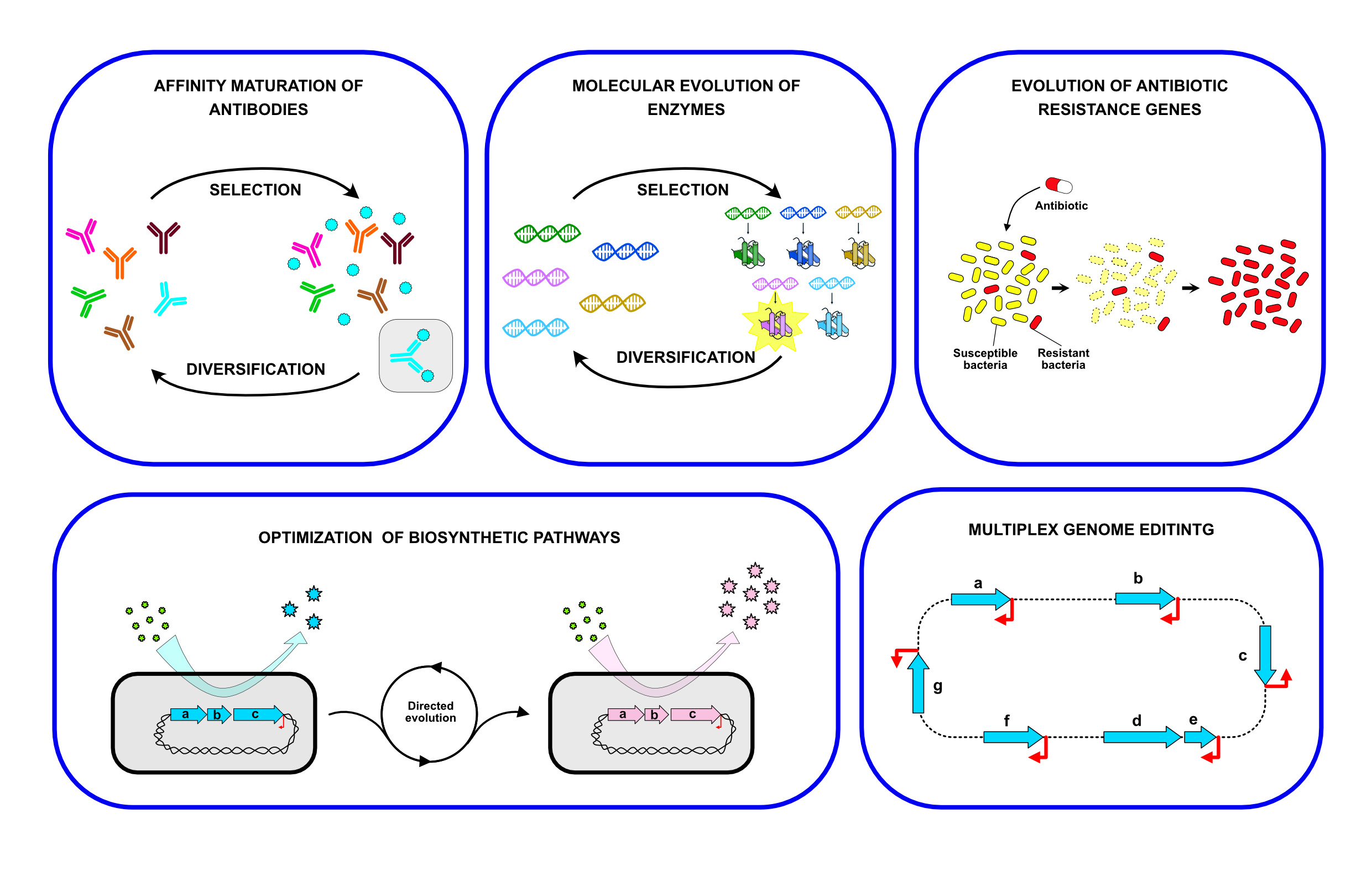 Figure 3. Potential applications of the T7-DIVA system
Figure 3. Potential applications of the T7-DIVA system
References
- Salema, V. and L. Fernández, Escherichia coli surface display for the selection of nanobodies. Microb Biotechnol, 2017. 10(6): p. 1468-1484.
- Ruano-Gallego, D., B. Álvarez, and L.Á. Fernández, Engineering the Controlled Assembly of Filamentous Injectisomes in E. coli K-12 for Protein Translocation into Mammalian Cells. ACS Synthetic Biology, 2015. 4(9): p. 1030-1041.
- Ruano-Gallego, D., et al., Screening and purification of nanobodies from E. coli culture supernatants using the hemolysin secretion system. Microbial Cell Factories, 2019. 18(1): p. 47.
- Lada, A.G., et al., Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry (Mosc), 2011. 76(1): p. 131-46.
- Komor, A.C., et al., Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 2016. 533(7603): p. 420-4.
- Nishida, K., et al., Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 2016. 353(6305).
- Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger, AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature, 2002. 418(6893): p. 99-103.
- Gaudelli, N.M., et al., Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 2017. 551: p. 464.
- Moore, C.L., L.J. Papa, 3rd, and M.D. Shoulders, A Processive Protein Chimera Introduces Mutations across Defined DNA Regions In Vivo. J Am Chem Soc, 2018. 140(37): p. 11560-11564.
- Bikard, D., et al., Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res, 2013. 41(15): p. 7429-37.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in