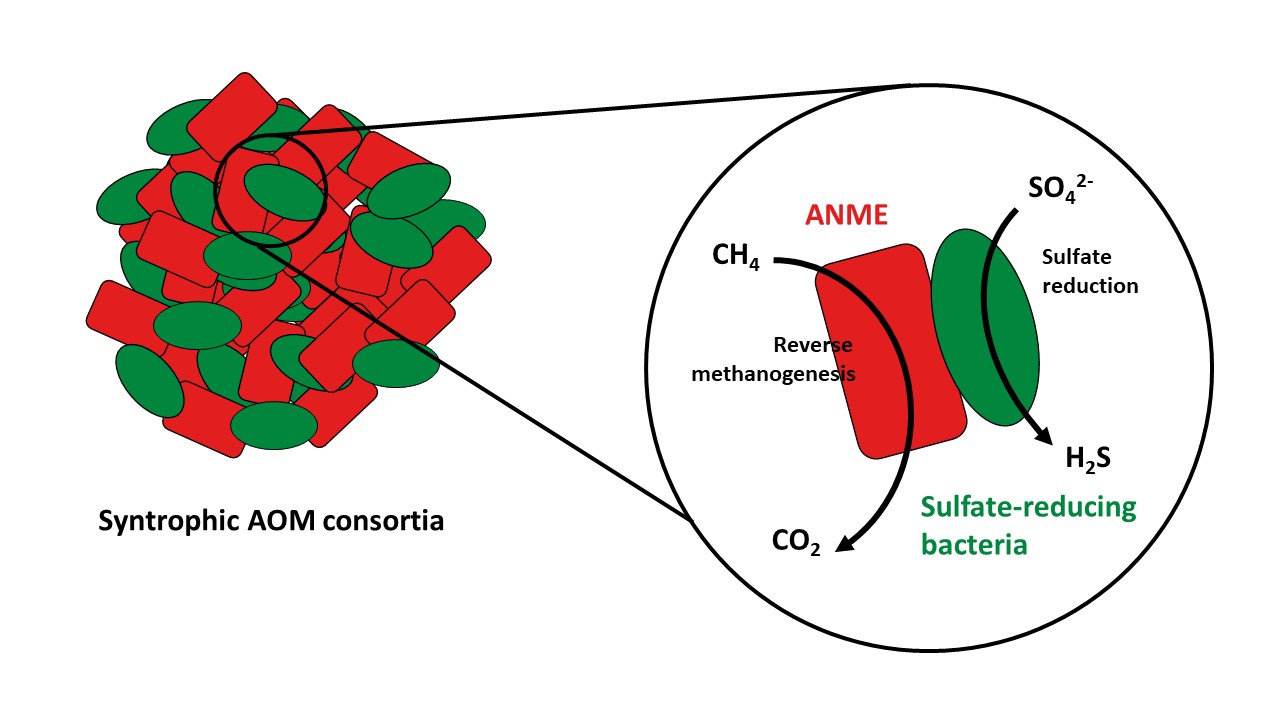
Methane is well-known as the second most important greenhouse gas. Fewer people know that the biggest producers of methane are not cows or rice fields, but deep-sea sediments. Luckily for us, microorganisms performing a metabolism called anaerobic oxidation of methane (AOM) prevent most of this methane produced in the deep-sea from entering the water column and eventually to the atmosphere. These organisms are known as ANaerobic MEthane-oxidizing archaea (ANME) and they were first described at the turn of the century 1-3. Since their discovery, researchers have identified different ANME groups (named ANME-1, ANME-2 and ANME-3) and have proposed a metabolic model of how these organisms degrade methane. In short, ANME oxidize methane to carbon dioxide by reversing the methanogenesis pathway. In the marine environment, ANME archaea have established syntrophic associations with sulfate-reducing bacteria, and the methane oxidation by ANME is coupled to sulfate reduction by bacteria. However, much remains unknown about the metabolism, ecology and function of ANME due to their difficulty in cultivation.
In 2019, fresh from my PhD, I took a postdoctoral position at MARUM (University of Bremen) under the supervision of Prof. Victoria Orphan. Victoria and her team at the California Institute of Technology have been studying the methane cycle for almost 20 years with a particular focus on AOM. She offered me a project to study the ANME communities of carbonate rocks from a recently discovered hydrothermal vent system, the South Pescadero Basin in the Gulf of California. Like many other hydrothermal sites, Pescadero looks like an alien world full of carbon spires that resemble the skyline of a futuristic city, amazing animals that survive in extreme conditions and vents that seem to be the gateway to the underworld. Since the discovery of Pescadero in 2017, Victoria and her team have visited the site three times on the research vessels Falkor and Nautilus operated by the Schmidt Ocean Institute and Ocean Exploration Trust respectively. They collected several sediment and rock samples to study the microbial diversity of this unique site. The sediment analysis by my colleague Daan Speth revealed a diverse thermophilic community 4. With some of the rock samples, my postdoc colleagues Antoine Crémière and Fabai Wu established different cultures unveiling a novel archaeal species called ‘Candidatus Heimdallarchaeum endolithica’ 5. Still many more rocks were waiting to reveal their secrets.

Although my new job was based in Bremen, I had to travel to the sunny California to start my research, as the carbonate samples were in Victoria’s lab in Pasadena. Here, with the help of Antoine I extracted DNA from variuous rocks to perform metagenomic analysis. Back in Bremen, my analysis showed that ANME-1 archaea were the dominant organisms in some of these rocks, which was not surprising given that these vents are a methane-rich environment. My initial goal was to find genomic features that would distinguish these carbonate-dwelling ANME-1 archaea from other ANME-1 present in the sediment and other rocks. Using bioinformatics, I tried to find a link between the phylogeny, the metabolism and the environment of these ANME-1, but inferring these relationships from genomes is sometimes tricky. After many analyses, in the middle of the COVID outbreak, I began to feel that I was at a dead end. I spoke to Victoria, and showed her my “limited results”: I had recovered several novel ANME-1 genomes from two groups (ANME-1a and an undescribed ANME-1 clade), but I could not link their genomic features to the carbonate environment. She was more optimistic than I was, and encouraged me to continue describing their metabolic potential.
At this point, I took a step back to summarize all the data and suddenly a new perspective emerged. In trying to find links between the ANME and the carbonates, I had overlooked the new ANME-1 clade, which we named ANME-1c. This clade is phylogenetically positioned at the base of the ANME-1 order, and the ANME-1c archaea are the closest relatives of the sister orders Syntrophoarchaeales and Alkanophagales, which cannot degrade methane, but can degrade larger alkanes. Indeed, the ANME-1c genomes encode distinct metabolic features that appear to be related to this deep-branching position. The most striking of these features was the presence of genes for a hydrogenase. Hydrogenases are enzymes that can either metabolize hydrogen or produce hydrogen. Since most ANME-1 genomes lack these genes, hydrogen has fallen out of favor as a molecular intermediate between the ANME and the syntrophic partner bacteria during AOM. Besides, the absence of hydrogenases suggests a strictly methanotrophic lifestyle, weakening the hypothesis that ANME-1 could act as a hydrogenotrophic methanogen. We therefore wondered what the role of these hydrogenases was in ANME-1c. Interestingly, the closest relatives of ANME-1, Syntrophoarchaeales and Alkanophagales, do encode for hydrogenases as well as a few ANME-1 genomes. Phylogenetic analysis revealed that hydrogenases appear to be an ancient trait of this entire group of organisms. Hydrogenase genes were vertically inherited by the common ancestor of all ANME-1 and then differentially lost within the different ANME-1 groups. Still, the role of these hydrogenases remains puzzling, since even within the ANME-1c groups, some genomes lack the corresponding genes, suggesting that they are part of a pangenomic repertoire rather than an absolute requirement for the ANME-1 function. Based on this information, we cannot predict the role of these hydrogenases, but a recent paper published during the review process of our paper showed that some ANME-1c do not express the hydrogenase genes during AOM6.
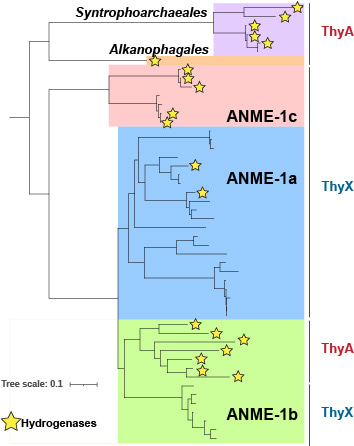
ANME-1c represent a novel clade at the base of the ANME-1
order. Stars indicate the presence of hydrogenase genes.
The presence of ThyX or ThyA is indicated on the right side
After these results, I started to draft a paper describing the ANME-1c clade in detail. At this point, my colleague Fabai contacted me because he had some ANME-1c genomes from the rock cultivation that might be interesting for the paper. In fact, one of them was the first closed ANME-1 genome. He also had some analysis of the virus targeting Pescadero ANME-1, which could give more information about these ANME-1. As he began to compile this new information, new findings emerged. We decided to publish together both stories as they cover different aspects of ANME-1 evolution. His analysis uncovered up to 16 undescribed virus families targeting ANME-1 archaea with a broad metabolic gene repertoire. Victoria, Fabai and I decided to discuss these new findings during a zoom call in three time zones spanning the whole globe (USA, Europe, China). During this call, Victoria pointed out that some viruses had genes for ThyX, an enzyme involved in the thymidine synthesis. This was exciting because ANME-1 are the only ANME group to use this enzyme, while the other ANME groups use another similar enzyme, ThyA 7. Similarly, the sister orders of Syntrophoarchaeales and Alkanophagales also use ThyA. When Fabai compared the ThyX of viruses and ANME-1, he discovered a shared origin, with the viral ThyX preceding those of ANME-1. Thus, it seems that the ANME-1 acquired the genes for thyX from viruses displacing the original thyA genes, which are still present in the sister orders.
To sum up, our results shed new light on the evolution of ANME-1 and its associated virome by combining genomics, metabolic potential and ecology. To honor the origin of the samples, we decided to name the new ANME-1c species after words of the local peoples of the Gulf of California (the Kiliwa and the Pa ipai) and the new viruses after Mayan gods.
References
1 Hinrichs, K.-U., Hayes, J. M., Sylva, S. P., Brewer, P. G. & DeLong, E. F. Methane-consuming archaebacteria in marine sediments. Nature 398, 802-805, doi:10.1038/19751 (1999).
2 Boetius, A. et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623-626, doi:10.1038/35036572 (2000).
3 Orphan, V. J. et al. Comparative Analysis of Methane-Oxidizing Archaea and Sulfate-Reducing Bacteria in Anoxic Marine Sediments. Applied and Environmental Microbiology 67, 1922-1934, doi:doi:10.1128/AEM.67.4.1922-1934.2001 (2001).
4 Speth, D. R. et al. Microbial communities of Auka hydrothermal sediments shed light on vent biogeography and the evolutionary history of thermophily. The ISME Journal 16, 1750-1764, doi:10.1038/s41396-022-01222-x (2022).
5 Wu, F. et al. Unique mobile elements and scalable gene flow at the prokaryote–eukaryote boundary revealed by circularized Asgard archaea genomes. Nature Microbiology 7, 200-212, doi:10.1038/s41564-021-01039-y (2022).
6 Benito Merino, D., Zehnle, H., Teske, A. & Wegener, G. Deep-branching ANME-1c archaea grow at the upper temperature limit of anaerobic oxidation of methane. Frontiers in Microbiology 13, doi:10.3389/fmicb.2022.988871 (2022).
7 Chadwick, G. L. et al. Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea. PLOS Biology 20, e3001508, doi:10.1371/journal.pbio.3001508 (2022).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in