The Great Escape
Published in Microbiology

The paper in Nature Microbiology is here: http://go.nature.com/2BJSofo
A few months into my PhD, I was giving a ten minute talk to my fellow peers to introduce my project on malaria parasite release (egress) from host red blood cells (RBCs) when a dreaded technical issue cropped up. A critical video showing the sequential morphology changes during egress would not play. In my mildly panicked attempt to describe the events in words, I slipped into my everyday vocabulary instead of more academic phrases: “First the parasitophorous vacuole membrane in which the parasite develops and multiplies ‘pops’ (=ruptures), which allows the parasites to ‘jiggle about more’ (=increased mobility), and finally the red blood cell goes ‘boom’ (=ruptures)!” While it was an unconventional description of the process, malaria parasite egress from the RBC has certainly been described as an explosive event. It is in part the destruction of the RBCs that leads to disease.
When the red blood cell goes boom - releasing more parasites to repeat the cycle of infection and destruction! *Thanks to James Thomas for the video.
Work on egress over the last 20 years or so had shown that proteases were important for the process and that there is sequential ‘popping’ of the parasitophorous vacuole membrane (PVM) followed by the RBC membrane (RBCM). However, the questions of how and what was causing the respective rupture events remained unanswered - until now. In our paper published in Nature Microbiology, we identify a parasite serine protease, SUB1 as being essential for the rupture of the PVM plus all the subsequent steps in egress. In contrast a cysteine protease, SERA6, which is activated by SUB1, is essential only for the final step in egress, rupture of the RBC membrane (RBCM).
But how did James Thomas (the co-first author) and I come to work on SUB1 and SERA6 in the first place? Well, previous work in the Blackman lab had shown that SUB1 was released from parasite secretory organelles into the PV just prior to egress and this secretion was regulated by a parasite protein kinase called PKG. We also actually knew quite a lot about SUB1 as an enzyme thanks to the successful recombinant expression of SUB1; we had a crystal structure, a list of known substrates and activity characteristics. However, the exact biological significance of SUB1 activity was a mystery as all this work was done in the ‘dark ages’ of genetic manipulation of the malaria parasite. With the available gene disruption technologies, directly knocking out essential genes resulted in no viable parasites, and no parasites meant no phenotype. Luckily, a new tool in the genetic manipulation kit was developed around the time James joined the lab. Now, we are able to generate robust conditional knockout mutants, so we can grow up parasites then rapidly knock out the genes of interest, allowing us to monitor and study the parasites at the precise point in their short developmental lifecycle at which they are arrested.
SERA6 had been previously identified as a SUB1 substrate, but recombinant expression of SERA6 has been (and remains) an insurmountable challenge, making further characterisation of SERA6 seem impossible. However, by approaching this problem from the perspective of the parasite, James uncovered the role of SERA6 in RBCM rupture. I was then assigned the mission of figuring out how SERA6 was mediating RBCM rupture - what does this protease cleave? A prime candidate from the get-go was the RBC cytoskeleton, a chicken wire-like protein mesh lying underneath the RBCM that provides structural strength. But the cytoskeleton is essentially a huge complex of multiple proteins, so which constituent was it? I recall an early project briefing session with Mike Blackman where, after studying together some of our early SDS PAGE gels he confidently stated: “I bet you its spectrin (the major component of the cytoskeleton, made up of alpha and beta subunits).” And indeed, the primary substrate of SERA6 does seem to be beta-spectrin!
Whilst we have figured out some parts of the choreography of egress, there are still many unanswered questions. How exactly does SUB1 mediate PVM rupture? How does SUB1 activate SERA6? Are there additional as yet undiscovered substrates of SERA6? These are only a small selection of the questions we are working towards answering. Nevertheless, we hope that we will eventually know enough about how the malaria parasite bursts out of the RBC to use that knowledge to develop new therapies against it.
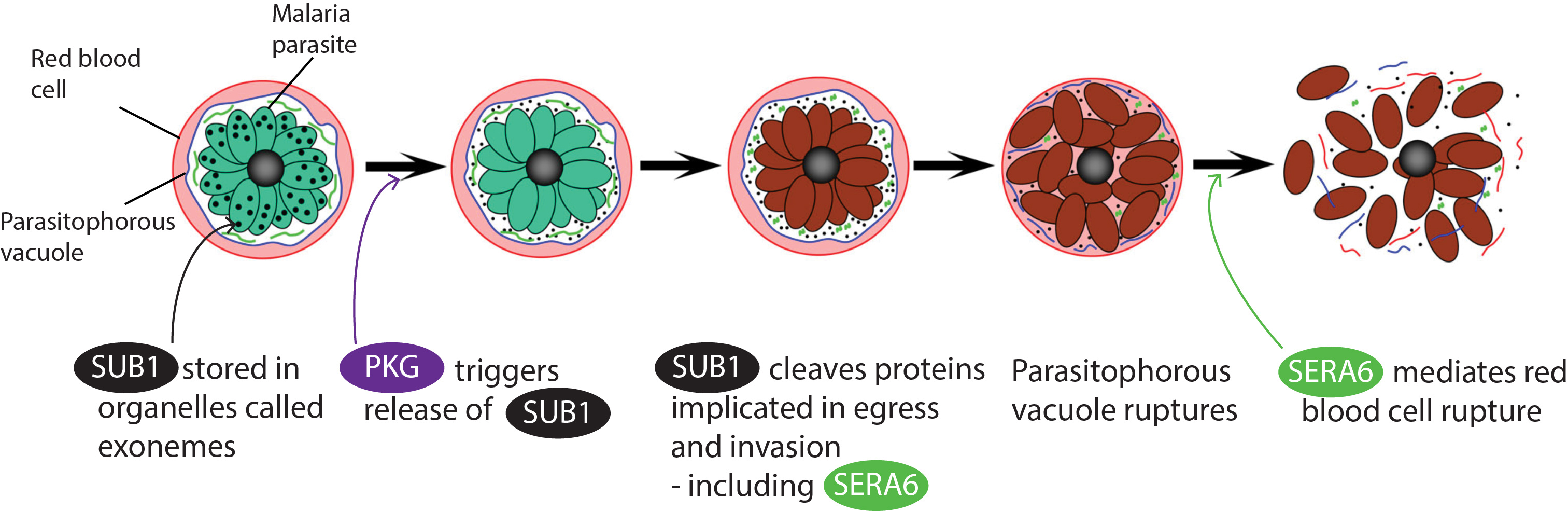
Egress choreography: a work in progress…

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Thank you Michele for a very nice summary, a great title and a cool video! And congratulations on the paper - to you and all the co-authors.
Love your alternative video description!
Boom! Great post.