The small plasmid that could
Published in Ecology & Evolution

Plasmids are agents of lateral gene transfer that facilitate rapid bacterial adaptation by transferring beneficial traits between microbial populations. Prominent examples are the transfer of genes encoding resistance mechanisms to antibiotics or pesticides. Under non-selective conditions plasmids are, however, not essential and are considered costly to the host cell. Nonetheless, plasmids are ubiquitous in nature where they constitute an integral part of microbial genomes. In our study, we supply a mechanistic explanation for how plasmids persist under non-selective conditions over evolutionary time scales.

Plasmids are drivers of adaptation to growth limiting conditions over a short time (e.g., in the presence of antibiotics1) as well as responsible for long-term transitions of life style modes (e.g., emergence of pathogenicity2). The most striking examples for the impact of plasmids on transfer of genes are documented in ecosystems exposed to fluctuating levels of antibiotics, for example in clinics or in industrial farming. Here, due to strong positive selection, antibiotic resistance is rapidly propagated in bacterial populations. Although many plasmids encode genes that can potentially benefit their bacterial hosts, very often plasmids were shown to be a metabolic burden on the individual host cell. Consequently, in the absence of selection for plasmid-encoded traits, plasmid-free cells outcompete plasmid-carrying cells and that can, potentially, drive to plasmid extinction. This discrepancy in plasmid biology where plasmids may provide a benefit and, at the same time impose a cost, has led to several theories and theoretical models to explain their long time persistence in natural microbial communities (e.g.,3).
The paradox of plasmid existence led to the questions that built the framework for my PhD project and our study. Why do we find so many diverse – and sometimes cryptic – plasmids in various bacterial species without apparent selection pressure was most puzzling and – at the same time – most exciting for us. Our initial view on the plasmid paradox was along the lines of the neutral theory4. What if plasmids could be neutral (‘invisible’) to their hosts? Our main objective was to study the forces that influence plasmid persistence over evolutionary timescales. In our project we were fortunate to combine expertise in the field of microbial evolution (our lab) with those of Prof. Itzhak (Itzik) Mizrahi (Ben-Gurion University, Israel; https://lifeserv.bgu.ac.il/wp/imizrahi/) whose group is studying the dynamics and ecology of plasmids in natural environments. A study published by Itzik’s group in 2012 unraveled the vast abundance and diversity of plasmids in the rumen microbiome while coining the term plasmidome5. The authors further found a correlation between plasmid content with their ecological niche, similarly to the known correlation of bacterial species composition with specific habitats. While discussing their interesting results during a visit in Itzik’s lab in Ben-Gurion, we realized that to be able to understand the abundance and dynamics of plasmids in natural ecosystems, fundamental questions regarding plasmid persistence and evolution needed to be addressed. Thus, going back to a model organism such as Escherichia coli in a controlled experimental setup was the next logical step.
Many studies focused their research on plasmids with a large genome size (>50 Kbp) that encode several accessory genes (e.g.,6). In contrast, small plasmids that don’t confer a clear function to their host remain largely understudied. These plasmids are, however, widespread in nature and might constitute an unknown source of evolutionary innovations (e.g., found in rumen plasmidome). Moreover, mechanisms that lead to their stable inheritance up to the degree of being a part of the bacterial lineage remain elusive.
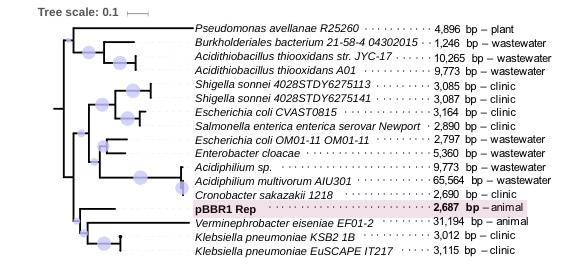
Therefore, we decided to conduct a long-term evolution experiment with a small model plasmid carrying a single resistance gene (pCON). Our model plasmid is of broad-host-range (pBBR1 origin) and widespread in nature. To follow the plasmid frequency over time (i.e., number of plasmid carrying cells) we chose the long-established method of replica plating developed by Joshua and Esther Lederberg7. Incidentally, Joshua Lederberg invented the term plasmid in 19528. A detail, which made us confident that we picked the fitting monitoring system for our experiments. Nonetheless, a factor that we didn’t expect was the difficulty to find cotton velvet, which is necessary to replicate the agar plates. Who would have thought that only kings’ velvet is suitable for E. coli populations?
Confident and highly motivated, I began the evolution experiments following a 12-hour transfer rhythm – 9 am and 9 pm. After a while – two months without interruption – the laborious transfer and plate replication system started to lead to a little decrease in motivation and simultaneously to a significant increase in coffee and chocolate consumption. However, I was rewarded; nothing compares to the enjoyment of making an exciting novel discovery. In my case, I observed the emergence of plasmid stability without selection for the plasmid presence (i.e., without antibiotics). I felt that all the experiment was worth the one mutant population I discovered. The evolved plasmid carried a 497 bp duplication between the origin of replication and the constitutively expressed antibiotic resistance gene. From this point all our discoveries slowly fell into place.
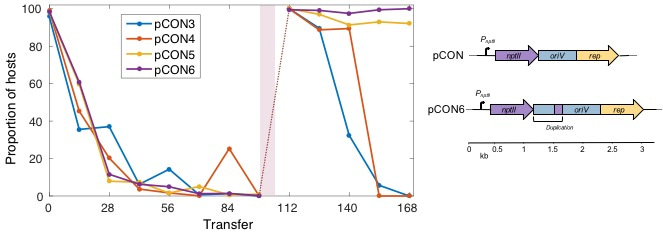
Plasmids are often considered costly to their host and are therefore expected to be eliminated from the lineage by purifying selection. We demonstrate that a small plasmid can be fitness neutral and stably inherited regardless of the selection pressure. We further find that plasmid stability may be determined by transcription-replication conflicts which can be alleviated by plasmid-intrinsic recombination as well as influenced by environmental conditions. Overall, the emergence of plasmid stability under non-selective conditions provides an evolutionary explanation for the ubiquity of plasmids – and the genes they carry – in nature.
References
1. San Millan, A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26, 978–985 (2018).
2. Bruto, M. et al. Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 11, 1043–1052 (2017).
3. Maclean, R. C. & San Millan, A. Microbial evolution: Towards resolving the plasmid paradox. Curr. Biol. 25, R764–R767 (2015).
4. Kimura, M. The neutral theory of molecular evolution. (1983). doi:10.1017/cbo9780511623486.010
5. Brown Kav, A. et al. Insights into the bovine rumen plasmidome. Proc. Natl. Acad. Sci. USA 109, 5452–5457 (2012).
6. Porse, A., Schönning, K., Munck, C. & Sommer, M. O. A. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873 (2016).
7. Lederberg, J. & Lederberg, M. E. Replica plating and indirect selection of bacterial mutants. J. Bacteriology 63, 399–406 (1952).
8. Lederberg, J. Cell genetics and hereditary symbiosis. Physiol. Rev. 32, 403–430 (1952).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in