Three RNA splicing-related genetic variants and lung cancer risk: Results from the TRICL-ILCCO and OncoArray projects
Published in Cancer

Aberrant RNA splicing has been implicated in the ancestry-related biology of cancer disparities and as a potential source of novel targets for precision oncology1. The role of aberrant splicing as a primary cause of Mendelian diseases has been widely accepted by decades of related studies2-4. However, much less has been reported and appreciated regarding the extent of physiological RNA splicing variation among human populations and the phenotypic variability and disease susceptibility affected by them in humans5.
Lung cancer is the most common malignancy in humans, leading the causes of cancer death worldwide6. Although a large number of germline mutations within cancer susceptibility genes have been reported, genetic etiology of lung cancer is still not fully understood7,8. In addition, until recently, despite the important role of RNA splicing in cancer, limited efforts have been made in the genome-wide profiling of RNA splicing-related variation in cancer patients, including in lung cancer9.
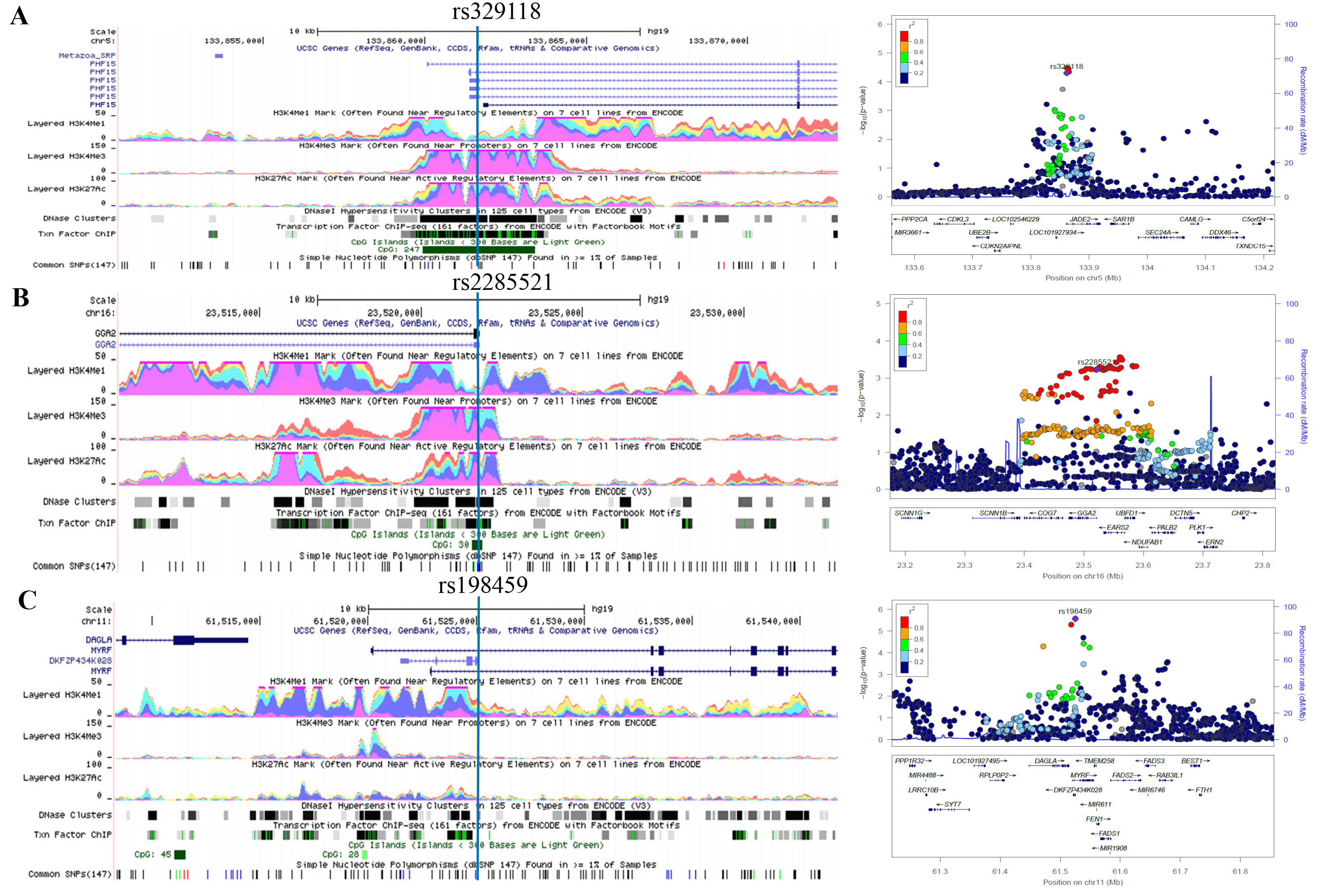
To conduct a comprehensive characterization of a genome-wide profile of common lung cancer genetic susceptibility loci associated with RNA splicing, we first performed a meta-analysis to discover potential RNA splicing-related SNPs using summary statistics from eight published lung cancer genome-wide association studies (GWASs) from the Transdisciplinary Research in Cancer of the Lung (TRICL) and the International Lung Cancer Consortium (ILCCO)10. Those significant SNPs discovered were then validated using data from the OncoArray platform that provides an unprecedented opportunity for additional de novo discovery as well as for fine mapping of lung cancer susceptibility7,11. For those identified SNPs that were found to be significant in both discovery and validation datasets, we further performed stratified analyses by smoking status and histological type and investigated their effects on gene expression and potential regulatory mechanisms in cell lines and tissues by using the available genomic and genetic data from multiple public databases.
In the present study, with the largest lung cancer study population of European ancestry ever reported, we identified three genetic variants in genome-wide profiling RNA splicing-related genes to be associated with lung cancer risk. We believed that all susceptibility alleles, if biologically meaningful, should be correlated with expression levels of the corresponding genes in normal lymphocytes and/or lung tissues.
References
- Al Abo, M. et al. Differential alternative RNA splicing and transcription events between tumors from African American and White patients in The Cancer Genome Atlas. Genomics 113, 1234-1246 (2021).
- Dong, X. & Chen, R. Understanding aberrant RNA splicing to facilitate cancer diagnosis and therapy. Oncogene 39, 2231-2242 (2020).
- Scotti, M.M. & Swanson, M.S. RNA mis-splicing in disease. Nat Rev Genet 17, 19-32 (2016).
- Boussaad, I. et al. A patient-based model of RNA mis-splicing uncovers treatment targets in Parkinson's disease. Sci. Transl. Med. 12, eaau3960 (2020).
- Park, E., Pan, Z., Zhang, Z., Lin, L. & Xing, Y. The Expanding Landscape of Alternative Splicing Variation in Human Populations. Am. J. Hum. Genet. 102, 11-26 (2018).
- Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 68, 394-424 (2018).
- McKay, J.D. et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 49, 1126-1132 (2017).
- Li, T., Kung, H.J., Mack, P.C. & Gandara, D.R. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J. Clin. Oncol. 31, 1039-1049 (2013).
- Li, Y. et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 393, 40-51 (2017).
- Liu, H. et al. Functional variants in DCAF4 associated with lung cancer risk in European populations. Carcinogenesis 38, 541-551 (2017).
- Amos, C.I. et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol. Biomarkers Prev. 26, 126-135 (2017).
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
Tumor-type-agnostic biomarkers and treatments in oncology
Publishing Model: Open Access
Deadline: Mar 05, 2026
Emerging adjuvant and neo-adjuvant treatment approaches in solid tumors
Publishing Model: Open Access
Deadline: Mar 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in