
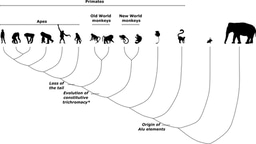
When it comes to human evolution, Africa rightly takes center stage. Fossil finds from East and South Africa document the emergence of the big-brained and bipedal Homo genus more than 2 million years ago1, while modern human presence outside Africa has been traced back using genomics to movement out of Africa more than 50,000 years ago2,3. But the discovery of extinct hominin fossils outside Africa – from Dmanisi hominins in Georgia4 to Dragon Man in China5 – have shown that parallel experiments in hominin evolution were taking place across neighboring Eurasia through much of the past 2 million years.
The recovery of DNA from fossils of extinct Eurasian hominins has revealed that distinct human species in Africa and Eurasia were not always evolving in mutual isolation. The first evidence of interhominin entanglement came through comparisons between Neanderthal and modern human genomes: individuals of European and Asian descent inherit a small fraction of their genomes (1-4%) from Neanderthals6. Modern humans emerging from Africa had met and mated with Neanderthals in Europe by about 40,000 years ago before populating the rest of the world.
In the years since this discovery, our view of recent human evolutionary history has been transformed, with many new chapters and indeed, new Eurasian hominins coming to light.
The European deficit
Dating of Neanderthal remains found in sites spanning much of western Eurasia suggests they had emerged by at least 140,000 years ago and disappeared about 40,000 years ago7,8. The recovery of the first complete Neanderthal nuclear genome from specimens in Vindija Cave in Croatia and the subsequent detection of Neanderthal DNA in the genomes of modern non-Africans resolved long-standing debates in the field regarding Neanderthal contribution to our gene pool6. But this was soon followed by the somewhat perplexing observation that East Asians appeared to have approximately 20% more Neanderthal ancestry than Europeans9, suggesting a more complex history of encounters between Neanderthals and modern humans than a single meeting when modern humans entered Eurasia about 50,000 years ago.
Early explanations for higher Neanderthal ancestry in East Asians centered on a second encounter with Neanderthals in Asia after splitting off from Europeans10,11. More recent methods do point to multiple encounters between Neanderthals and both East Asian and European populations, some of which may partially explain the European deficit in Neanderthal ancestry12,13. However, a strong accumulating body of evidence suggests that a single major encounter between Neanderthals and modern humans may yet be consistent with what we observe in modern genomes.
In 2016, Lazaridis and colleagues showed that individuals living during and before the Neolithic in the Near East carried substantial genomic ancestry from a basal Eurasian group containing no appreciable Neanderthal ancestry14. This basal group likely separated from other non-Africans – who would go on to mate with Neanderthals – soon after leaving Africa. Early Near Eastern farmers carrying the basal Eurasian ancestry migrated into Europe, diluting some of the Neanderthal admixture in contemporary European populations by mixing with them.
Coll Macià and colleagues recently showed that Neanderthal fragments found in East Asians and Europeans cover substantially overlapping parts of the genome and most likely derive from a single encounter with Neanderthals15. Why, then, do Neanderthal fragments in East Asians tend to be longer than in Europeans? The authors provide evidence that the fragments in East Asians tend to be longer as a result of longer generation intervals. Recombination between parental chromosomes breaks apart adjacent genomic regions and will have progressively decreased the length of Neanderthal fragments in human genomes over time; Europeans have shorter fragments because they went through more generations.
Finally, Chen and colleagues found that Africans also contain some Neanderthal ancestry, likely from a back-migration of Europeans containing Neanderthal ancestry into Africa16. The Neanderthal fragments uniquely shared between Europeans and Africans were previously not interpreted as Neanderthal, leading to underestimation of Neanderthal ancestry in Europeans.
Even while assuming a single major encounter between Neanderthals and early non-African humans, the 20% European deficit in Neanderthal ancestry may thus be largely explained by a combination of shorter generation interval compared to East Asians, back-migration of Europeans into Africa, and dilution of Neanderthal ancestry by mixing with a basal Eurasian population that never or minimally interbred with Neanderthals.
Chen and colleagues also found that some of the sequences shared between Neanderthals and Africans had passed into Neanderthals via an earlier human migration out of Africa between 100,000 and 150,000 years ago, long before they mated with Eurasians about 40,000-50,000 years ago.
The lost mitochondrial genome
To examine more evidence of earlier human migrations out of Africa, we must meet another extinct Eurasian hominin group, the Denisovans, whose genomes live on as fragments in modern Pacific Islanders, and to a lesser extent, in East, South, and Southeast Asians17. Denisovans were first discovered through sequencing DNA from a single hominin finger bone excavated from Denisova cave in Siberia18. The Denisovan nuclear genome places it closer to Neanderthals than to modern humans, with the common ancestor of Denisovans and Neanderthals likely diverging from the ancestor of modern humans more than 550,000 years ago19.

Neanderthal mitochondrial DNA – which is distinct from nuclear genomic DNA and passed down maternally – presented an early paradox. While the Neanderthal and modern human nuclear genomes diverged more than 550,000 years ago, their mitochondrial genomes appeared to have diverged about 430,000 years ago. Comparisons with Denisovan mitochondrial DNA cemented the contradiction: Neanderthals were more similar to modern humans in their mitochondrial DNA than to their closer evolutionary group, the Denisovans.
The paradox is explained by an early migration of a group ancestral to modern humans out of Africa into Europe followed by interbreeding with Neanderthals and complete replacement of their mitochondrial DNA. The divergence time of all known Neanderthal mitochondrial genomes is about 270,000 years ago19. The introduction of the African mitochondrial lineage into Neanderthal ancestors must have occurred sometime between 270,000 and 430,000 years ago.
A 2020 study provided further evidence of an earlier African movement into Europe by finding DNA deriving from ancient African humans in the Neanderthal nuclear genome, that was introduced into ancestral Neanderthals between 200,000 and 300,000 years ago20. It seems plausible that this is the same event that led to the mitochondrial genome replacement, which would indicate that the human fragments in the Neanderthal genome came from an ancient group of humans that had split off from our African lineage by about 430,000 years.
Combined with Chen and colleagues’ finding that there was possibly ancient human-Neanderthal interbreeding 100,000-150,000 years ago, this suggests the possibility of multiple Out-of-Africa forays into Europe. Neanderthals kept records of these movements in their genomes, pointing to a more mobile and connected world of human species during the Middle Paleolithic than previously known.
The not-quite Neanderthals
New hominin fossils from the Sima de los Huesos site in Spain, first reported in 2014, were immediately thought to represent Neanderthal ancestors21. Dated to about 430,000 years ago, their teeth and bones share many features with Neanderthals, but lower cranial capacity and primitive nasal features suggested that they weren’t quite Neanderthals.
The recovery of nuclear and mitochondrial DNA from the Sima de los Huesos hominins – the oldest hominin DNA extracted to date – presented the opportunity to place the morphological inferences under a genomic lens. Meyer and colleagues found that these early Eurasian hominins did indeed represent ancestral Neanderthals that had already split off from Denisovans22, which is consistent with the older dates for the Neanderthal-Denisovan split predicted by previous studies.
A mitochondrial genome recovered from these hominins was more closely related to Denisovans than to Neanderthals. This suggests that Meyer and colleagues had caught the Neanderthal ancestors before their mitochondrial genomes were replaced by encounters with ancient humans from Africa, neatly bookending a chapter of early human history that we could not have learnt about without the genomes of our hominin cousins from Eurasia.
New threads
The presence of Neanderthal and Denisovan ancestry in many modern human populations raises the possibility that there are other as yet undiscovered extinct hominins we may be harboring genes from. And indeed, novel approaches for detecting archaic hominin fragments in genomes without knowing the identity of the hominins themselves are beginning to provide tantalizing clues to other dispersals and interactions between hominins across Eurasia, in addition to those involving Neanderthals and Denisovans23. For instance, Hubisz and colleagues recently discovered that 1% of the Denisovan genome derives from an unknown archaic hominin and 15% of these fragments made it into modern humans following interbreeding with Denisovans in Eurasia20. It is tempting to speculate that this unknown hominin was in fact a species known from the fossil record, such as Homo erectus, but without the genome of the hominin itself, we cannot know for certain.
A decade since the first complete Neanderthal genome sequence was reported, interbreeding between contemporary hominin species has become well-established at least for the timespan in which DNA can be instructive. There is little reason to believe this was not true throughout the entire history of the hominin lineage.
Bibliography
- Dunsworth, H. M. Origin of the Genus Homo. Evol. Educ. Outreach 3, 353–366 (2010).
- Scheinfeldt, L. B., Soi, S. & Tishkoff, S. A. Working toward a synthesis of archaeological, linguistic, and genetic data for inferring African population history. Proc. Natl. Acad. Sci. 107, 8931 (2010).
- Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016).
- Ferring, R. et al. Earliest human occupations at Dmanisi (Georgian Caucasus) dated to 1.85-1.78 Ma. Proc. Natl. Acad. Sci. U. S. A. 108, 10432–10436 (2011).
- Ji, Q., Wu, W., Ji, Y., Li, Q. & Ni, X. Late Middle Pleistocene Harbin cranium represents a new Homo species. The Innovation 2, 100132 (2021).
- Green Richard E. et al. A Draft Sequence of the Neandertal Genome. Science 328, 710–722 (2010).
- Devièse, T. et al. Direct dating of Neanderthal remains from the site of Vindija Cave and implications for the Middle to Upper Paleolithic transition. Proc. Natl. Acad. Sci. 114, 10606 (2017).
- Douka, K. et al. Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova Cave. Nature 565, 640–644 (2019).
- Wall, J. D. et al. Higher Levels of Neanderthal Ancestry in East Asians than in Europeans. Genetics 194, 199 (2013).
- Kim, B. Y. & Lohmueller, K. E. Selection and Reduced Population Size Cannot Explain Higher Amounts of Neandertal Ancestry in East Asian than in European Human Populations. Am. J. Hum. Genet. 96, 454–461 (2015).
- Vernot, B. & Akey, J. M. Complex History of Admixture between Modern Humans and Neandertals. Am. J. Hum. Genet. 96, 448–453 (2015).
- Villanea, F. A. & Schraiber, J. G. Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol. 3, 39–44 (2019).
- Taskent, O., Lin, Y. L., Patramanis, I., Pavlidis, P. & Gokcumen, O. Analysis of Haplotypic Variation and Deletion Polymorphisms Point to Multiple Archaic Introgression Events, Including from Altai Neanderthal Lineage. Genetics 215, 497–509 (2020).
- Lazaridis, I. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016).
- Coll Macià, M., Skov, L., Peter, B. M. & Schierup, M. H. Different historical generation intervals in human populations inferred from Neanderthal fragment lengths and mutation signatures. Nat. Commun. 12, 5317 (2021).
- Chen, L., Wolf, A. B., Fu, W., Li, L. & Akey, J. M. Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals. Cell 180, 677-687.e16 (2020).
- Vernot Benjamin et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352, 235–239 (2016).
- Reich, D. et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010).
- Posth, C. et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nat. Commun. 8, 16046 (2017).
- Hubisz, M. J., Williams, A. L. & Siepel, A. Mapping gene flow between ancient hominins through demography-aware inference of the ancestral recombination graph. PLOS Genet. 16, e1008895 (2020).
- Arsuaga J. L. et al. Neandertal roots: Cranial and chronological evidence from Sima de los Huesos. Science 344, 1358–1363 (2014).
- Meyer, M. et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507 (2016).
- Teixeira, J. C. & Cooper, A. Using hominin introgression to trace modern human dispersals. Proc. Natl. Acad. Sci. 116, 15327 (2019).



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in