Uncovering distinct genomic and molecular signatures in triple-negative inflammatory breast cancer
Published in Cancer
The Need for Better Diagnostic Markers for Triple-Negative Inflammatory Breast Cancer
Inflammatory Breast Cancer (IBC) is a rare but aggressive form of breast cancer that spreads rapidly. While it represents only 2-4% of breast cancer cases, IBC is responsible for a disproportionate number of breast cancer-related deaths. Despite advances in chemotherapy, surgery, and radiation, the five-year survival rate for IBC remains alarmingly low at just 40%, compared to about 90% for other types of breast cancer.
Currently, IBC is diagnosed primarily based on physical symptoms like redness, swelling, and a thickened, dimpled skin texture on the breast, often described as an "orange peel" appearance. However, relying on these symptoms alone can lead to delays in diagnoses, contributing to poorer outcomes.
Triple-negative inflammatory breast cancer (TN-IBC), a particularly challenging subtype of IBC, lacks hormone receptors (ER, PR) and HER2 protein, limiting available treatment options. This subtype is associated with poorer prognosis, underscoring the urgent need for better diagnostic and therapeutic strategies.
To better understand this aggressive cancer, our team conducted the most comprehensive study to date, analyzing the genetic, transcriptomic, and immune characteristics of TN-IBC tumors from patients enrolled in a clinical trial and comparing them to non-TN-IBC tumors from another protocol. All samples were without treatment exposure at the time of collection to ensure unbiased insights.
Key Findings:
Genetic, Transcriptomic, and Immune Differences in TN-IBC
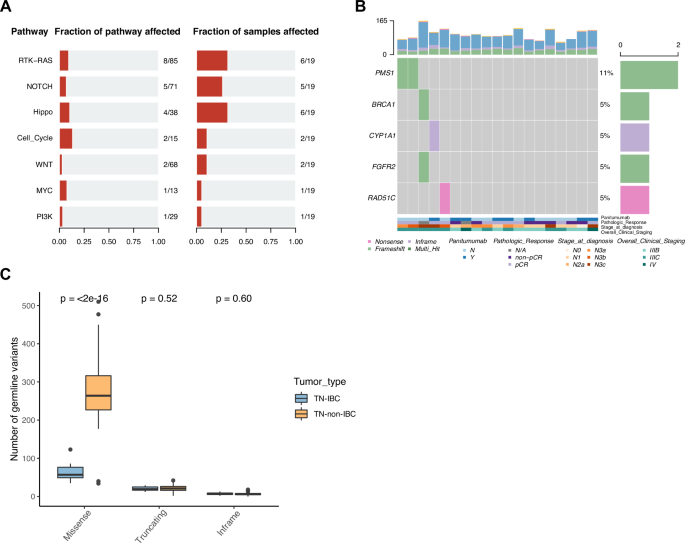
The study revealed that TN-IBC tumors had fewer mutations than triple-negative breast cancer (TNBC). However, several genes exhibited higher rates of alteration in TN-IBC, including GATA3, PIK3CA, ARNT, DDR2, BCL9, FCGR2B, and LMNA. Notable, ARNT, DDR2, BCL9, FCGR2B, and LMNA are being reported in IBC research for the first time, opening new avenues for investigation.
We also identified proteins and microRNAs uniquely overexpressed in TN-IBC tumors compared to TNBC. PIK3CA, in particular, stood out, as it has high amplification alteration and expression in TN-IBC tumors, making it a promising target for future treatments. Additionally, we found higher expression of FGA, FGB, and FGG genes, which encode the polypeptide chains of the protein fibrinogen, in TN-IBC, suggesting fibrinogen may play a role in the growth and progression of TN-IBC tumors.
Moreover, our study revealed that TN-IBC tumors have an immunosuppressive microenvironment, meaning they can evade the immune system more effectively than other types of breast cancer. This may be one of the reasons why TN-IBC is particularly aggressive.
Understanding Treatment Responses
Our study also looked at how TN-IBC patients responded to treatments, including chemotherapy alone and chemotherapy combined with an antibody targeting the EGFR protein.
We found that patients who responded better to chemotherapy tended to have more gene mutations than those who didn’t. This trend was also observed with the chemotherapy combined with the EGFR-targeted treatment, although more samples are needed to confirm this finding. This suggests that certain genetic changes might help predict whether a patient will benefit from a treatment.
We identified several genes that might be potential targets to improve TN-IBC’s response to these treatments. Additionally, immune cells linked to poorer treatment response were identified in the tumor microenvironment. Targeting these immune cells could be a new avenue for treatment.
What This Means for the Future
This research has provided valuable insights into TN-IBC, revealing its unique genetic, transcriptomic, and immune characteristics that could lead to improved diagnosis and treatment options. By identifying specific markers and genetic traits, we aim to develop better diagnostic tools and therapies that increase survival rates for patients facing this difficult type of breast cancer.
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Ask the Editor – Inflammation, Metastasis, Cancer Microenvironment and Tumour Immunology
Got a question for the editor about inflammation, metastasis, or tumour immunology? Ask it here!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
AI Approaches in Drug Design
Publishing Model: Open Access
Deadline: Mar 31, 2026
Genomic Instability
Publishing Model: Open Access
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in