Understanding the RNA virosphere at a landscape scale
Published in Microbiology

RNA viruses likely infect all cellular organisms in diverse environments. Despite their expanding diversity with the application of high-throughput sequencing technology in virus discovery, our knowledge of the ecology and evolution of RNA viruses remains limited, due to major sampling biases, historical dependence on cell culture and PCR for finding viruses, and a focus on those pathogenic to humans or economically important animals and plants. More importantly, the following questions remain vague: What is the role of viruses in biosphere in addition to the etiologic agents of humans and animals and plants? How many viruses are circulating in nature? What factors determine the diversity and composition of viruses in a specific environment?
The idea of this study could date back to the spring of 2015, the time when we discovered unprecedented diversity of RNA viruses in arthropods and revealed that arthropods are central to virus evolution. Also at that time, our work on invertebrate RNA virosphere was halfway done. These works were finally published in eLife and Nature in 2015 and 2016, respectively. Based on the findings of largely expanded phylogenetic and genomic diversity of RNA viruses in these studies, I began to consider: now that there is such a huge diversity of RNA viruses in invertebrates, how are their diversity and composition in natural environments (soil, water, sediment, etc.) – which are the “home” of various organisms including prokaryotes, invertebrates and vertebrates, even mammals? What role are they playing in a local ecosystem?
With these questions, immediately, I arranged two PhD students, Yong-Chao Li (jointly trained with Professor Run-Lin Ma at Institute of Genetics and Developmental Biology of Chinese Academy of Sciences) and Jian-Wei Shao (jointly trained with Professor Jei Yan at School of Medicine of Zhejiang University School of Medicine), who just joined my team, to conduct the investigation. In May of 2015, I took them to collect soil and sediment samples around China CDC in Beijing, and performed pre-experiments. In 2016, we also collected some samples in Henan and Heilongjiang provinces. However, the study progressed very slowly due to the shortage of qualified persons in my lab.
The turning point came with the arrival of a newcomer in 2017. My good friend Professor Zhong-Dao Wu at Zhongshan School of Medicine of Sun Yat-Sen University sent a postdoc Dr. Yan-Mei Chen to my lab as an exchange visitor. Chen has some experience in environmental microbiology, and more importantly, she is clever and hardworking! She immediately joined in field survey with another member of my team - Dr. Xiao Chen from South China Agricultural University - in Guangdong and Guangxi provinces, and she then came to my lab in Beijing with all the samples in August, 2017. During September 2017 to June 2018, we went to Wuhan and surrounding regions of Hubei province; Yili, Boertala and Hami of Xinjiang Uygur Autonomous Region; Yancheng of Jiangsu province; Neixiang, Lushi and Jiyuan of Henan province; Wenzhou of Zhejiang province; Hainan province; Yunnan province; Sichuan province; the Inner Mongolia Autonomous Region and Heilongjiang province, and collected samples from various environments, as well as feces of domestic animals. These sampling sites were selected to represent the major geographic locations and ecotypes in China as many as possible. My team members and co-operators (Jun-Hua Tian, Xiao Chen, Xian-Dan Lin, Zhong-Yu Hao, Zhuo-Cheng Zhou, Jin-Jin Shen, Hao Chen, Jian-Bo Wang, Ming Li etc.) all had done what they could in the field sampling. In addition, this research also involved the ocean ecosystem. In May and August of 2018, Dr. Xiao Chen went to South China Sea twice for sampling. In particular, we obtained the rare and precious deep-sea samples across South China Sea from Dr. De-Xiang Wang of Xiamen University. Dr. Wang suffered with cancer and knew that he might not have enough time to continue his studies. He contacted Dr. Xiao Chen and donated the samples he collected to us.

In addition to sampling, Yan-Mei spent more time in lab works in 2018. Based on the goal of the project, she improved the methods and finished most of the experiments with the help of several other students in my lab in Beijing. In my initial idea, Yan-Mei might be more suitable to be in charge of another project based on her expertise. However, due to the personnel changes in my lab, she had to take on the soil virome project. In order to speed up the process of data analysis, my good friend and also my long-term cooperator Professor Edward C. Holmes at Sydney University arranged his PhD student Sabrina Sadiq to join in the project, and sent Sabrina to China for one month in 2019. Sabrina is very talented and her participation no doubt greatly accelerated the process of the study! In the early January of 2020, we began to perform the experiment of virus confirmation in my lab in Beijing, which was eventually finished by Yan-Mei and several MSc or PhD students including Wen-Hua Gao, Xiu-Qi Luo, Jin-Jin Guo, etc. by the end of June 2020.
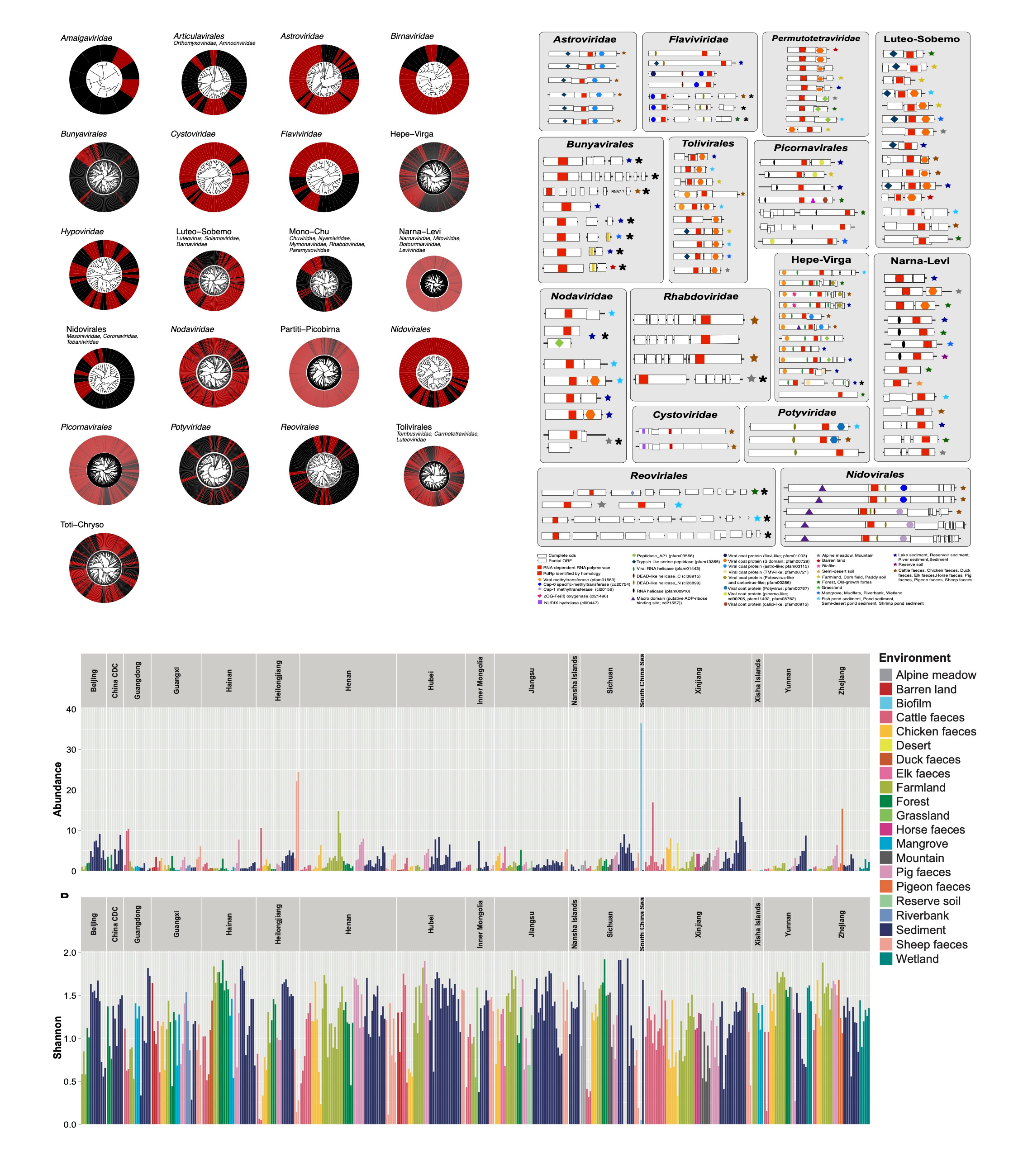
As mentioned above, we collected soil, sediment, and faeces from wild and livestock animals from 32 different geographic and physical environments covering 14 provinces and regions (including a wide range of sea areas in South China Sea) across China, and investigated the total RNA virome by metatranscriptomics (i.e., RNA-sequencing). From this, we identified 6,624 putatively novel virus operational taxonomic units (vOTUs) from at least 62 viral families, greatly expanding known diversity of the RNA virosphere. Sediments and animal faeces were particularly rich sources of viruses. Virome compositions were affected by local environmental factors including organic content and eukaryotic species abundance. Notably, environmental factors had a greater impact on the abundance and diversity of plant, fungal, and bacterial viruses than on animal viromes, suggesting that the former have adapted to their specific ecotypes. These data reveal the diversity of viruses at a landscape scale and suggest that they are an integral part of environmental ecosystems. Finally, our data also indicate that the diversity and composition of environmental RNA viruses is shaped by hosts and habitats.
 There were a lot of stories during the course of the study. When we collected samples in Wenzhou in 2017, I accidentally fell into a five-meter-deep ravine of a mountain, resulted in my three ribs and one vertebra broken, and had to stay in the hospital for some days. During that period, it was my co-operator Xian-Dan Lin that took over and continued the field surveys in Wenzhou, with the help of Yan-Mei and other students. What is unforgettable is that they came to the hospital to take care of me when they finished the sampling every day. I am very grateful for their kindly care and helps. In addition, my team had undergone various difficulties during the period, and considerable changes after the COVID-19 outbreak. Nevertheless, the majority of my teammates kept on the research and never gave up. I would like to express my sincerest appreciation to all the people who constantly support and care for me and my team. It’s their continued supports and concerns that kept us going through our science works.
There were a lot of stories during the course of the study. When we collected samples in Wenzhou in 2017, I accidentally fell into a five-meter-deep ravine of a mountain, resulted in my three ribs and one vertebra broken, and had to stay in the hospital for some days. During that period, it was my co-operator Xian-Dan Lin that took over and continued the field surveys in Wenzhou, with the help of Yan-Mei and other students. What is unforgettable is that they came to the hospital to take care of me when they finished the sampling every day. I am very grateful for their kindly care and helps. In addition, my team had undergone various difficulties during the period, and considerable changes after the COVID-19 outbreak. Nevertheless, the majority of my teammates kept on the research and never gave up. I would like to express my sincerest appreciation to all the people who constantly support and care for me and my team. It’s their continued supports and concerns that kept us going through our science works.

After working on this study for seven years, I am very excited that our work has now been published in Nature Microbiology. The completion of this study depends on the joint efforts of all the people involved, including the lab team and the field team. Many thanks to each member of my team who have stuck together through the years, against all odds, especially to the selfless behind-the-scenes contributors who were not mentioned in the paper. We really appreciated Dr. De-Xiang Wang for selflessly donating the precious samples to us and giving full play to the scientific value of these samples. I think he would be happy to see that in heaven.
Finally, also most importantly, I am very grateful to my wife Bin-Xiu Zhao, who worked at China CDC as well, for her decades of support. Despite suffering from a severe illness, she had never complained about my long working hours, and instead had done whatever she could for me. Unfortunately, she passed away in October of 2019, ……
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in