Unlocking the Puzzle of Phage Proteins: Building Blocks for Fighting Antibiotic Resistance
Published in Chemistry, Ecology & Evolution, and Microbiology

In the world of microbes, evolution can take some surprising twists. In the classical view of evolution, gene is often considered the most fundamental unit of selection. It turns out, however, that bacterial viruses – phages - have a remarkable propensity to shuffle fragments of their genes which can help phages overcome bacterial resistance mechanisms. These findings suggest that viral evolution may sometimes be better understood from the point of view of protein domains rather than entire proteins.
Both phages and bacteria are known to frequently pick up new genes during interactions with other microbes or mobile genetic elements like viruses and plasmids. This phenomenon is known as horizontal gene transfer (HGT), and it's responsible for the astonishing diversity seen in prokaryote genomes.
So, here is the big question:
Does such horizontal transfer work only at the level of entire genes? There are some known examples of proteins that have modular structures i.e. are composed of fragments that potentially can be used in multiple combinations. They include proteins involved in recognizing bacterial hosts (receptor binding proteins, RBPs) and enzymes responsible for breaking down the bacterial cell wall (endolysins).
This phenomenon has practical implications. It has been harnessed in biotechnology, such as the search for new endolysins with antibacterial properties and the design of synthetic phages with altered host ranges.
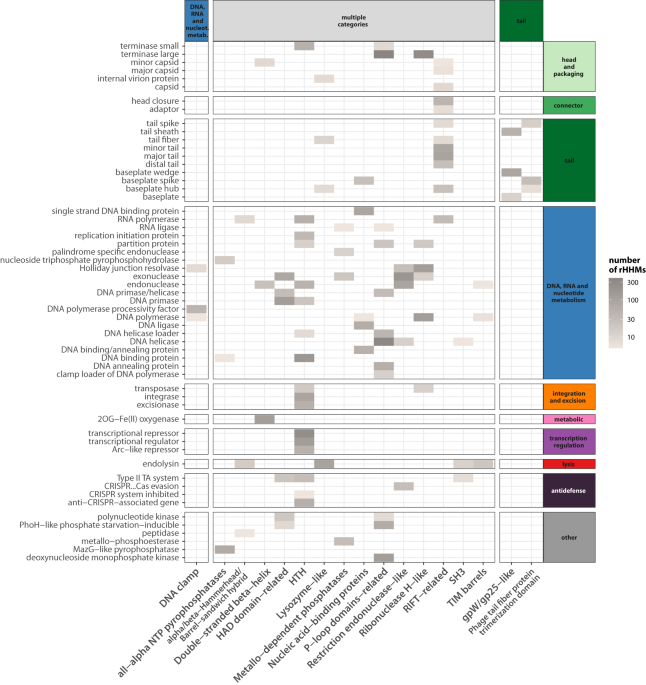
But is such modularity limited to the few examples reported so far in the literature? And what about other phage proteins? Can they also be composed of gene fragments that are reshuffled? To answer this question, we defined 'domain mosaicism.' This term describes pairs of proteins that share a similar fragment despite being otherwise unrelated.
When we examined thousands of phage genomes, we found that domain mosaicism is widespread in phage proteins of various functions, not limited to RBPs or endolysins. Moreover, it is not only present within proteins that have the same function. Similar fragments are also shared by proteins that perform different biological functions.
When we compared the extend of mosaicism between various phage proteins we found that the ones that are most mosaic apart from RBPs (i.e. tail fibers and tail spikes) and endolysins, are DNA polymerases.
Interestingly, not all phages have their own DNA polymerases, as some rely on their host's replication machinery. However, having their own DNA polymerases gives phages an edge in the evolutionary arms race, as it allows for more specific replication of their DNA and competition with the host's DNA polymerase. Finally, it allows to overcome multiple bacterial mechanisms that defend against phages.
In fact, the most mosaic proteins identified in this study are heavily involved in the ongoing battle between bacteria and phages and are often targeted by bacterial defense mechanisms. Therefore, they are under intense evolutionary pressure to diversify while maintaining their functions, and domain mosaicism is a key strategy to achieve this.
What is even more interesting, we found that the phenomenon of reshuffling domains within phage proteins is an on-going process and we showed a number of examples of such recent reshuffling within: tail fibres, endolysins but also DNA polymerases, replication initiation proteins, ribonucleotide reductases and neck passage proteins.
Why is it important?
In an era of rising antibiotic resistance, scientists are exploring new ways to combat bacterial infections, including using bacteriophages. Understanding the fundamental building blocks of phage proteins and how they function will help us design synthetic phages with tailored abilities or specific host ranges. Additionally, the knowledge of how domains can be shuffled and diversified, powered by new AI-aided technologiescan inform the design of modular phage-based tools, like biosensors, delivery vectors, and enzybiotics, all of which contribute to our global search for strategies to fight antibiotic resistance.
Moreover, the discovery of recent diversification via domain shuffling opens up possibilities for directed evolution experiments, which could lead to the creation of new phages with custom functionalities and host ranges.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in