Unraveling the complex evolution of color patterns
Published in Ecology & Evolution

Evolutionary studies of Heliconius butterflies often began with expeditions in the Amazon dating back more than 150 years. During that time, Bates and Müller described with wonder the variation in Heliconius wing patterns as if they were produced ‘‘by touch of an enchanter’s wand’’ (comments of Bates in Müller 1879, p. xxix). While Heliconius research has a deep root in evolutionary biology, there has been a recent wave of genetic discoveries underlying warning color diversity in Heliconius.

Figure 1. Sample of Heliconius erato diversity explored in the study.
Traditional crossing experiments in Heliconius beginning as early as the 1950’s started to describe a mix of over 30 linked and unlinked color pattern loci, including several that appeared to control major changes in wing coloration (Turner 1972; Sheppard et al. 1985). However, it wasn’t until 2005 that the genomic locations of these major color pattern loci began to be resolved (Tobler et al. 2005; Kapan et al. 2006; Kronforst et al. 2006; Joron et al. 2006; Counterman et al. 2010; Baxter et al. 2010). Most exciting was that the major color pattern loci described in several Heliconius species mapped to the same genomic regions (Joron et al. 2006). Genetic mapping quickly collapsed the more than 30 previously described color pattern loci to only a handful of genomic loci. This made what once seemed complex, to actually be controlled by a simple genetic “toolkit” (Reed et al. 2011; Martin et al. 2012; Nadeau et al. 2016).
In a massive collaboration, including ten institutions from seven countries, we obtained Heliconius samples from most of its widespread range throughout Central and South America. With this dataset in hand, a number of diverse bioinformaticians were lured to continue dissecting the complex genetic architecture of wing pattern variation.

Figure 2. Sample of Heliconius researchers who contributed to the study. From top left to bottom right: Steven Van Belleghem, Pasi Rastas, Alexie Papanicolaou, Simon Martin, Carlos Arias, Megan Supple, Joseph Hanly, James Mallet, James Lewis (with Larry Gilbert), Heather Hines, Mayte Ruiz, Camilo Salazar, Mauricio Linares, Gilson Moreira (picture missing), Chris Jiggins, Owen McMillan, Brian Counterman and Riccardo Papa.
The collaborative efforts resulted in the assembly of a new
reference genome for Heliconius erato
with the best current lepidopteran genome statistics to date. This reference
and the comparison of genomes among the diverse set of H. erato samples
generated a new comprehensive image of the genetic architecture underlying
diversity in Heliconius.
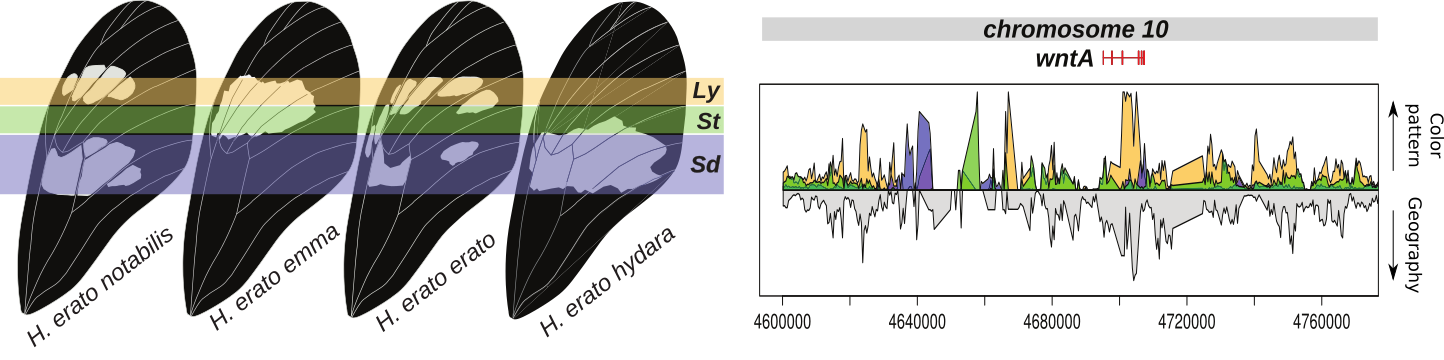
Figure 3. Example of identification of regulatory regions near the wntA gene that determine the shape of the forewing band. Graph on the right indicate regions in the genome that show high support for phylogenetic clustering according to color pattern rather than geography.
With this study, we are excited to make Heliconius
color pattern genetics complex once again! We find that at the major color pattern loci, there are much smaller regulatory regions
that modulate color pattern variation in a fashion that provides a flexible
mechanism for rapid diversification of color patterns (Van Belleghem et al. 2016). However, we are perhaps
most intrigued by the predictive ability of our academic forefathers that
hypothesized our genomic results from patterns of phenotypic variation.
Frederik Nijhout, for one, pioneered how genes may generate specific patterns
and boundaries in butterfly wings and sketched us a prototype “ground-plan” for
butterfly wing patterns (Nijhout 1991). Complementary to this ground-plan, Larry
Gilbert had long discussed how a simple set of genetic switches that control
the opening and closing of windows and shutters across the butterfly wing could
explain nearly all the color pattern variation (Gilbert 2003).
At Heliconius meetings his presentations
often involved over-sized cut outs of wings with various “window cutouts” in
the black wing, and various colored and sized “shutters” that could be placed
over the windows. We believe we have now identified the genomic regions that
regulate many of those window and shutter switches. We have also shown that
although there is a simple toolkit of color patterning genes involved,
underneath each gene lies a complexity of regulatory switches.
References
Baxter S. W., Nadeau N. J., Maroja L. S., Wilkinson P., Counterman B. a, Dawson A., Beltran M., Perez-Espona S., Chamberlain N., Ferguson L., Clark R., Davidson C., Glithero R., Mallet J., McMillan W. O., Kronforst M., Joron M., Ffrench-Constant R. H., Jiggins C. D., 2010 Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in the Heliconius melpomene clade. PLoS Genet. 6: e1000794.
Counterman B. a, Araujo-Perez F., Hines H. M., Baxter S. W., Morrison C. M., Lindstrom D. P., Papa R., Ferguson L., Joron M., Ffrench-Constant R. H., Smith C. P., Nielsen D. M., Chen R., Jiggins C. D., Reed R. D., Halder G., Mallet J., McMillan W. O., 2010 Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in Heliconius erato. PLoS Genet. 6: e1000796.
Gilbert L. E., 2003 Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic “‘tool box’” from synthetic hybrid zones and a theory of diversification. In: Boggs CL, Watt WB, Ehrlich PR (Eds.), Ecology and Evolution Taking Flight: Butterflies as Model Systems, University of Chicago Press, Chicago, US, pp. 1–34.
Joron M., Papa R., Beltrán M., Chamberlain N., Mavárez J., Baxter S., Abanto M., Bermingham E., Humphray S. J., Rogers J., Beasley H., Barlow K., ffrench-Constant R. H., Mallet J., McMillan W. O., Jiggins C. D., 2006 A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 4: e303.
Kapan D. D., Flanagan N. S., Tobler A., Papa R., Reed R. D., Gonzalez J. A., Restrepo M. R., Martinez L., Maldonado K., Ritschoff C., Heckel D. G., McMillan W. O., 2006 Localization of müllerian mimicry genes on a dense linkage map of Heliconius erato. Genetics 173: 735–757.
Kronforst M. R., Kapan D. D., Gilbert L. E., 2006 Parallel genetic architecture of parallel adaptive radiations in mimetic Heliconius butterflies. Genetics 174: 535–539.
Martin A., Papa R., Nadeau N. J., Hill R. I., Counterman B. A., Halder G., Jiggins C. D., Kronforst M. R., Long A. D., McMillan W. O., Reed R. D., 2012 Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. 109: 12632–12637.
Nadeau N., Pardo-Diaz C., Whibley A., Supple M., Saenko S. V., Wallbank R. W. R., Wu G. C., Maroja L., Ferguson L., Hanly J. J., Hines H., Salazar C., Dowling A., Ffrench-Constant R. H., Llaurens V., Joron, M., McMillan W. O., Jiggins C. D., 2016 The gene cortex controls mimicry and crypsis in butterflies and moths. Nature: 106–110.
Nijhout H. F., 1991 The Development and Evolution of Butterfly Wing Patterns. Smithsonian Institution Press.
Reed R. D., Papa R., Martin A., Hinas H. M., Counterman B. A., Pard-Diaz C., Jiggins C. D., Chamberlain N. L., Kronforst M. R., Chen R., Nijhout H. F., McMillan W. O., 2011 optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 333: 1137–1141.
Sheppard P. M., Turner J. R. G., Brown K. S., Benson W. W., Singer M. C., 1985 Genetics and the Evolution of Muellerian Mimicry in Heliconius Butterflies. Philos. Trans. R. Soc. B Biol. Sci. 308: 433–610.
Tobler A., Kapan D., Flanagan N. S., Gonzalez C., Peterson E., Jiggins C. D., Johntson J. S., 2005 First-generation linkage map of the warningly colored butterfly Heliconius erato. Heredity. 94: 408–417.
Turner J. R. G., 1972 The genetics of Some polymorhic form of the butterflies Heliconius melpomene (Linnaeus) and Heliconius erato (Linnaeus). II. The hybridization of subspecies of H. melpomene from Surinam and Trinidad. Zoologica 56: 125–157.
Van Belleghem S.M., Rastas P., Papanicolaou A., Martin S.H., Arias C.F., Supple M.A., Hanly J.J., Mallet J., Lewis J.J., Hines H.M., Ruiz M., Salazar C., Linares M., Moreira G.R.P., Jiggins C.D., Counterman B.A., McMillan W.O. & Papa R., 2017 Complex modular architecture around a simple toolkit of wing pattern genes. Nature Ecology & Evolution 1:0052.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in