Determining the molecular details of heme acquisition in Acinetobacter baumannii was a fortuitous outcome of my investigation into lipoprotein biology. The Moraes lab specializes in the study of bacterial membrane transport systems, particularly an outer membrane lipoprotein translocon discovered by our lab called Slam [1]. First identified in the meningitis causative pathogen, Neisseria meningitidis, Slams were later discovered throughout the Gram-negative proteobacteria by a former Graduate student, Yogesh Hooda [2]. Acinetobacter baumannii was a hit organism and piqued our interests for several reasons. Considered a hot pathogen, A. baumannii garnered significant media attention for infecting American soldiers in Iraq and Afghanistan. It also ranks as a top priority pathogen by the WHO and CDC for its extensive antibiotic resistance. A. baumannii Slam is found in a gene cluster that prior to the start of this study was indirectly linked to free heme utilization, a phenomenon largely unexplored in this organism. Slam is adjacent to a hypothetical protein (HphA) that we hypothesized to be a lipoprotein that functions in heme uptake and is dependent on Slam for cell surface localization. This was plausible as HphA contains a lipidation motif found in surface lipoproteins, and Slams are frequently found adjacent to the substrates they translocate. To further investigate the nature of Slams, the types of substrates they translocate, and shed light on a potential novel heme uptake mechanism, I decided to further characterize this Slam-HphA pair.
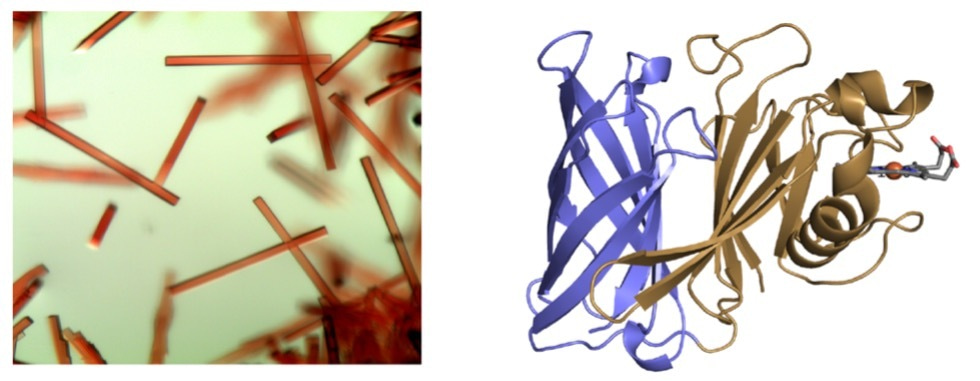
Being a structural biology lab, the natural first step towards understanding the function of HphA was to express, purify and crystallize the protein. Fortunately, HphA expressed well in E. coli and was straightforward to purify. We were excited by the deep red color of the protein, reflected in the beautiful rod-like crystals and indicative of bound heme (Figure 1a). We collected X-ray diffraction data remotely from the synchrotron at the Canadian Light Source. Luckily, we were able to obtain phase information and solve HphA’s structure using the anomalous signal from heme-iron, which saved time from pursuing other methods like heavy atom soaks and laborious Seleno-Met purifications. Heme was indeed bound to HphA through two His residues found in a clamp-like domain connected to an 8-stranded C-terminal β-barrel (Figure 1b). We discovered that HphA adopts a structure remarkably similar to Slam dependent surface lipoproteins.
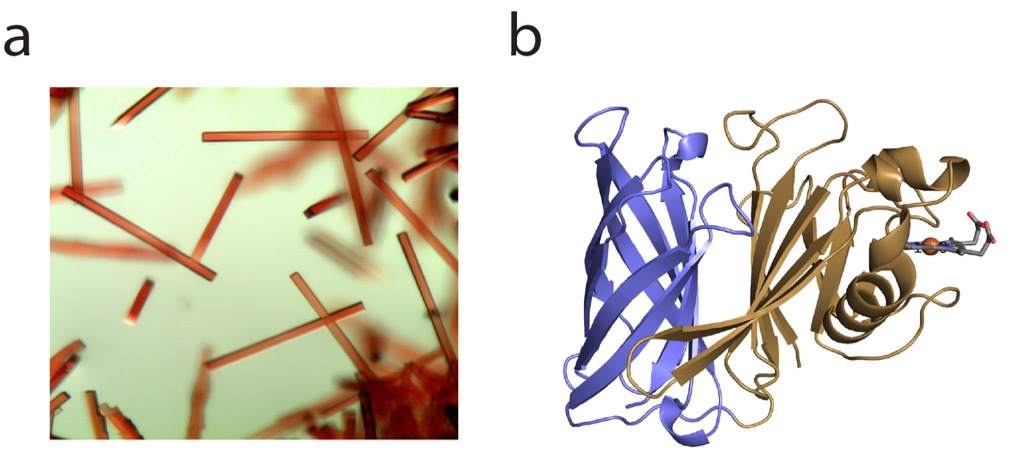
Figure 1: Crystals (a) and X-ray structure (b) of HphA (PDB 7RED). Heme is shown in stick form bound to the N-terminus.
Our lab had previously developed an E. coli based platform for testing a hypothetical lipoprotein’s dependence on Slam for cell surface exposure. Using this assay, we confirmed that HphA is dependent on Slam for localization to the surface of E. coli. Contrary to our results, a study published during my PhD convincingly showed that HphA is present in the media as opposed to being anchored on the surface of A. baumannii. Shifting from this artificial system to studying the native organism led to the discovery that Slam is required for HphA secretion into the environment. Slams functioning in general secretion was an exciting, controversial finding as up until this point, Slam was only known to be involved in surface lipoprotein translocation.
Structure-based functional inferences guided the rest of our in vitro work. Through pull-downs and spectroscopic assays, we showed that secreted HphA acts as a hemophore, capable of binding to and passively scavenging heme from the largest heme reservoir in humans, hemoglobin. To perform these assays, we had to search through the literature to find a way to strip HphA of its co-purifying heme. Although a harsh and long process, acidic acetone extraction proved successful. One downside of this was that HphA lost its cool color and we could no longer visually track the protein through purification columns. However, we were able to crystallize and solve the structure of apo (empty) protein by molecular replacement using holo (heme loaded) HphA, and gained important insights into the structural transitions that occur upon heme binding and release.
In addition to protein work, we investigated the biological importance of the Slam gene cluster to A. baumannii’s growth and virulence. The Acinetobacter community was incredibly instrumental in expediting our research and making the process as smooth as possible. We acquired transposon mutant strains and A. baumannii expression vectors from the Manoil lab at the University of Washington, and Visca lab at the University Roma Tre. These tools were important for establishing that the Slam gene cluster is essential for growth on human hemoproteins as sole iron/heme sources. Another important discovery was that an outer membrane protein in the Slam gene cluster referred to as HphR serves as the receptor for HphA, as heme would need to be released from the hemophore and transferred to a channel for entry into the cell.
The growth assays were nicely complemented with mice models of A. baumannii infection thanks to a long-standing collaborator, the Gray-Owen lab at the University of Toronto, and a new partnership with Dr. Wangxue Chen at the National Research Council of Canada in Ottawa. Together, these infection models demonstrated that Slam and its substrate are required for full virulence, but are not essential factors. On the other hand, HphR is absolutely essential for virulence and extrapulmonary spread of the bacteria from the lungs to the vasculature and distal organs. Based on our findings, we propose that HphR acquires host heme independently, however, HphA secreted into the environment by Slam enhances the efficiency of heme piracy and therefore, maximizes virulence (Figure 2).
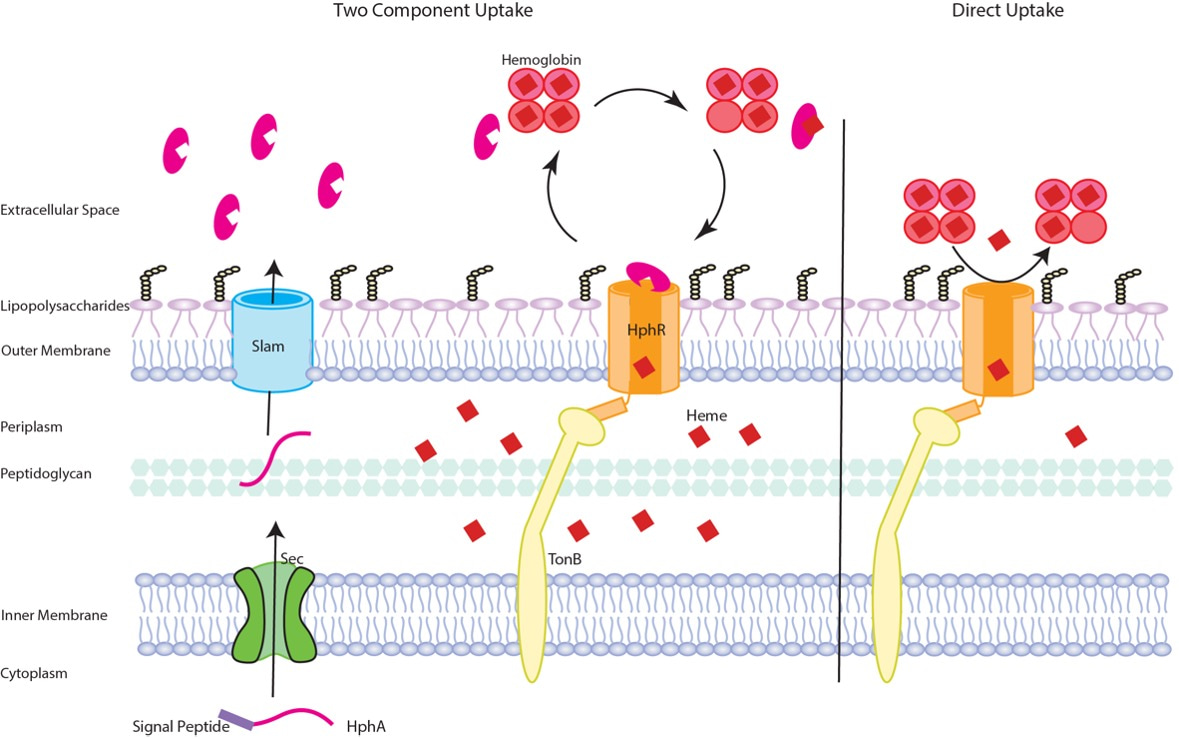
Figure 2: Schematic of heme acquisition by the Slam gene cluster. Briefly, HphA is secreted into the environment by Slam, where it pirates host heme and delivers it to the TonB dependent receptor, HphR, for transport across the outer membrane and into the cell. Alternatively, HphR acquires heme on its own, although this process is less efficient. Details can be found in the article linked below.
Check out the full article here:
https://www.nature.com/articles/s41467-021-26545-9.
References
[1] Hooda, Y. et al. Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nat Microbiol 1, 16009 (2016).
[2] Hooda, Y., Lai, C. C. L. & Moraes, T. F. Identification of a Large Family of Slam-Dependent Surface Lipoproteins in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 7, (2017).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in