Unveiling Subspecies-Level Differentiation in Acidithiobacillus ferrooxidans: Insights from Genomic Taxonomy and Resequencing analyses
Published in Microbiology

The field of microbiology has been revolutionized by advances in genomic taxonomy, redefining much of the tree of life, and allowing researchers to uncover hidden diversity within microbial populations and communities from most environments. The Acidithiobacillia class is no exception; recent studies utilizing genomic taxonomy methods have shed new light on this class of bacteria, revealing previously unrecognized genera and species, and contributing to clarifying the phylogenetic landscape of the class (1). However, this research has provided evidence of ongoing differentiation processes within certain species, highlighting the importance of understanding the factors driving such divergence. In this blog post, we share the stories behind some critical aspects discussed in our Scientific Reports paper.
Who is Acidithiobacillus ferrooxidans, and why should we care about knowing?
Our study focused on Acidithiobacillus ferrooxidans, a chemolithotrophic acidophile known for obtaining energy from the oxidation of inorganic molecules found in mineral ores; thus, we tag them as “rock eaters”. Strains of this species are metabolically versatile, using iron, sulfur, and hydrogen for biosynthesis and energy conservation via aerobic and anaerobic respiration (2, 3). These traits enable A. ferrooxidans representatives to thrive in environments that are uninhabitable to most, both for the scarcity of nutrients and the extremity of their pH (Fig. 1). One such habitat is found in man-made mining operations, where A. ferrooxidans is relevant in accelerating mineral dissolution and recovering valuable metals and critical elements. Therefore, A. ferrooxidans physiology, genetics and diversity are subjects of interest from basic and applied interests.
 Figure 1. Iron- and sulfur-oxidizing A. ferrooxidans in natural environments, liquid cultures, and on the plates.
Figure 1. Iron- and sulfur-oxidizing A. ferrooxidans in natural environments, liquid cultures, and on the plates.
Unveiling subspecies-level taxa and processes driving long-term differentiation.
In recent years many iron-oxidizing acidithiobacilli originally classified as A. ferrooxidans-like isolates have been reassigned into several new species based on cumulative evidence ranging from physiological to genomic (Fig. 2). Delving deeper into the genomic blueprints of these strains, we found evidence for additional species (candidate species `A. ferruginosus´) and subspecies-level taxa, showcasing uncovered mechanisms of long-term differentiation. We provide compelling evidence for two distinct sublineages within A. ferrooxidans by utilizing various criteria and parameters, including genomic relatedness metrics, synteny levels, gene content differences, and assessing the repertoire of mobile genetic elements (MGEs). These sublineages exhibit differential MGE repertoires, and MGE-associated gene cargo, encoding functions such as energy metabolism, ion transport, cell surface modification, and defense mechanisms. In the linked paper we present and discuss the traits and processes behind subspecies-level differentiation.
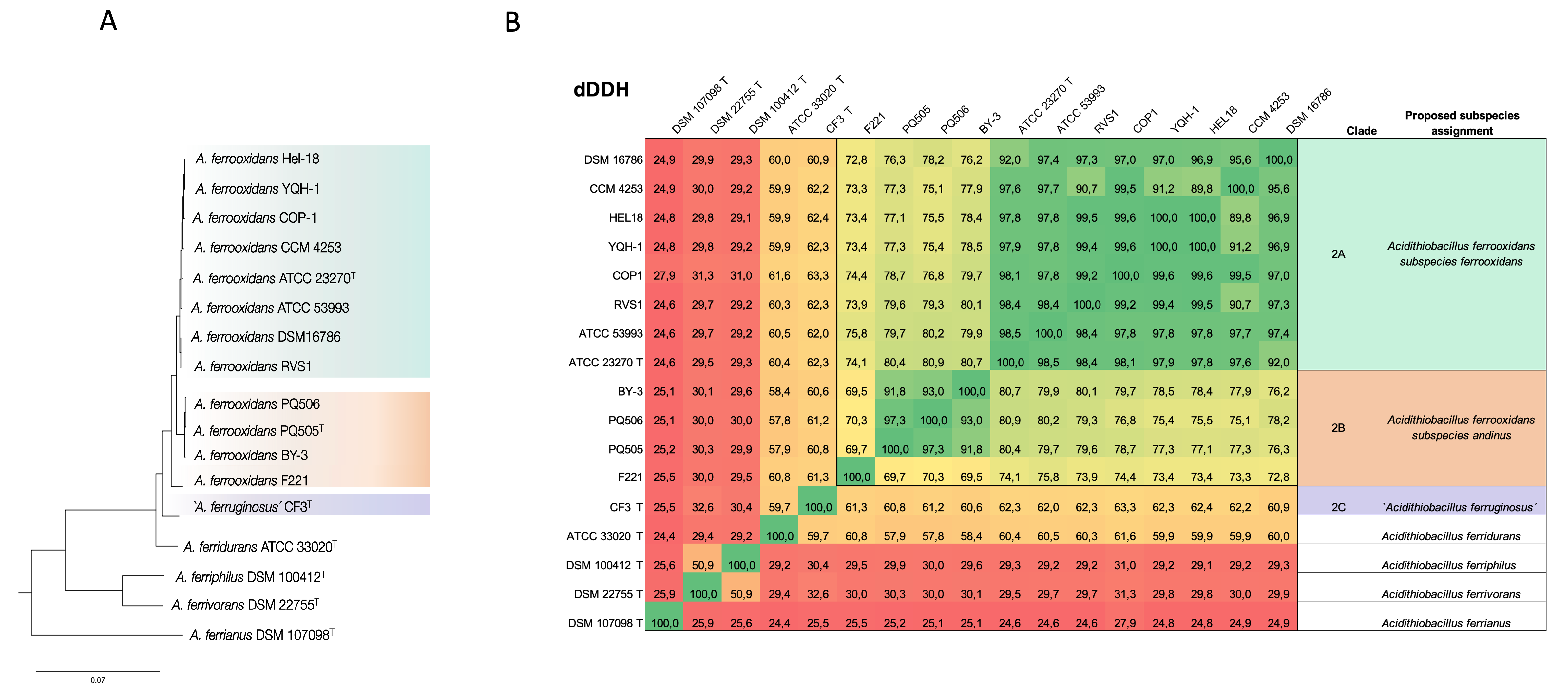
Figure 2. Phylogenetic tree and genome relatedness metrics delimit iron-oxidizing spp. and subspecies. (A) Core gene complement-based Maximum-Likelihood tree based on (1). (B) Genome relatedness based on digital DNA-DNA hybridization values. A. ferrooxidans sublineages 2A and 2B demonstrated distinct differences in synteny, further supporting the subspecies-level classification.
Resequencing experiments underpin a role for ISs in short-term adaptation.
Using iron- and sulfur-adapted cultures of sublineage 2A strain CCM 4253 as a test case and repeated transfer experiments to fresh culture medium containing either energy source, we aimed to investigate the impact of integrated MGEs (iMGEs) and episomal MGEs (eMGEs) on short-term adaptive processes (Fig. 3A). We thus monitored changes in genotype and phenotype when switching culture media in short-term adaptation experiments. Genome resequencing did not show significant changes in the location of iMGEs, nor the relative abundance of eMGEs. Instead, a single replicative transposition event of an active insertion sequence was detected in sulfur-adapted cultures, which impaired the ability of the bacterium to oxidize ferrous iron upon the switch of electron donors. The explanation behind this phenotype was traced to the pstC2 gene, the inactivation of which affected the ability of A. ferrooxidans cells to regulate phosphate transport by the pstII operon tightly, leading to cytoplasmic acidification. Consequently, this pH alteration caused a loss of the ability to oxidize ferrous iron, a crucial energy source for the bacterium. Reversion of the pstC2 mutation coincided with a shortened iron-oxidation lag phase, indicating the essential role of the pstC gene in energy metabolism during the oxidation of ferrous iron (Fig. 3B).
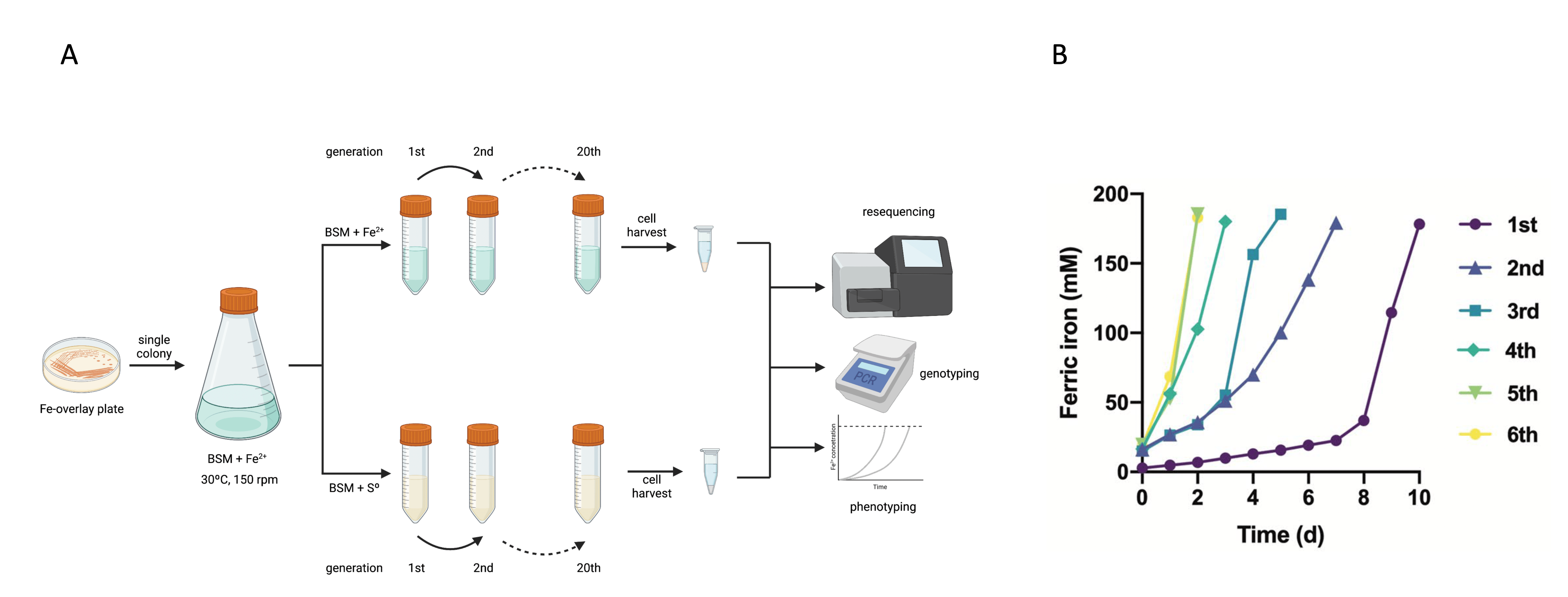
Figure 3. Effect of switching between electron donors in MGE-mediated adaptation of A. ferrooxidans sublineage 2A strain CCM 4253. (A) The experimental design. (B) Sulfur-adapted A. ferrooxidans mutant strain DpstC2 passaged on ferrous iron repeatedly.
The study on Acidithiobacillus ferrooxidans showcased the power of genomic taxonomy in unraveling subspecies-level differentiation. By analyzing the genetic characteristics of sequenced strains, we successfully identified two distinct sublineages within A. ferrooxidans, shedding light on their divergence and the underlying processes driving adaptation. The presence of specific mobile genetic elements, such as plasmids and integrated MGEs, played a significant role in the observed differentiation, influencing long-term adaptive processes. In turn, active insertion sequences profoundly impact short-term adaptive strategies. These findings provide a deeper understanding of the genetic mechanisms that enable A. ferrooxidans to adapt to changing growth conditions and open new avenues for further research in microbial evolution and biotechnology.
References:
- Moya-Beltrán A, Beard S, Rojas-Villalobos C, Issotta F, Gallardo Y, Ulloa R, Giaveno A, Degli Esposti M, Johnson DB, Quatrini R. Genomic evolution of the class Acidithiobacillia: deep-branching Proteobacteria living in extreme acidic conditions. ISME J. 2021 Nov;15(11):3221-3238. doi: 10.1038/s41396-021-00995-x.
- Quatrini R, Johnson DB. Acidithiobacillus ferrooxidans. Trends Microbiol. 2019 Mar;27(3):282-283. doi: 10.1016/j.tim.2018.11.009.
- Kucera J, Lochman J, Bouchal P, Pakostova E, Mikulasek K, Hedrich S, Janiczek O, Mandl M, Johnson DB. A Model of Aerobic and Anaerobic Metabolism of Hydrogen in the Extremophile Acidithiobacillus ferrooxidans. Front Microbiol. 2020 Nov 30;11:610836. doi: 10.3389/fmicb.2020.610836.
Follow the Topic
-
Scientific Reports

An open access journal publishing original research from across all areas of the natural sciences, psychology, medicine and engineering.
Related Collections
With Collections, you can get published faster and increase your visibility.
Obesity
Publishing Model: Hybrid
Deadline: Apr 24, 2026
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in