
Explore the Research

A Quantum-Chemical Bonding Database for Solid-State Materials - Scientific Data
Scientific Data - A Quantum-Chemical Bonding Database for Solid-State Materials
Understanding the physical and chemical phenomena that define the properties of solid-state materials has long been a quest at the heart of materials science. From a chemist's perspective, the intricate web of chemical bonds could play a major role in dictating material properties. In our paper "A Quantum Chemical Bonding Database for Solid State Materials," we dive into the world of chemical bonding that is aimed to uncover relationships between material properties and chemical bonding on a larger scale. Through a comprehensive bonding analysis of over 1,500 insulators and semiconductors, we have created a dataset that should be invaluable for researchers and scientists working on materials design, computational chemistry, and machine learning.
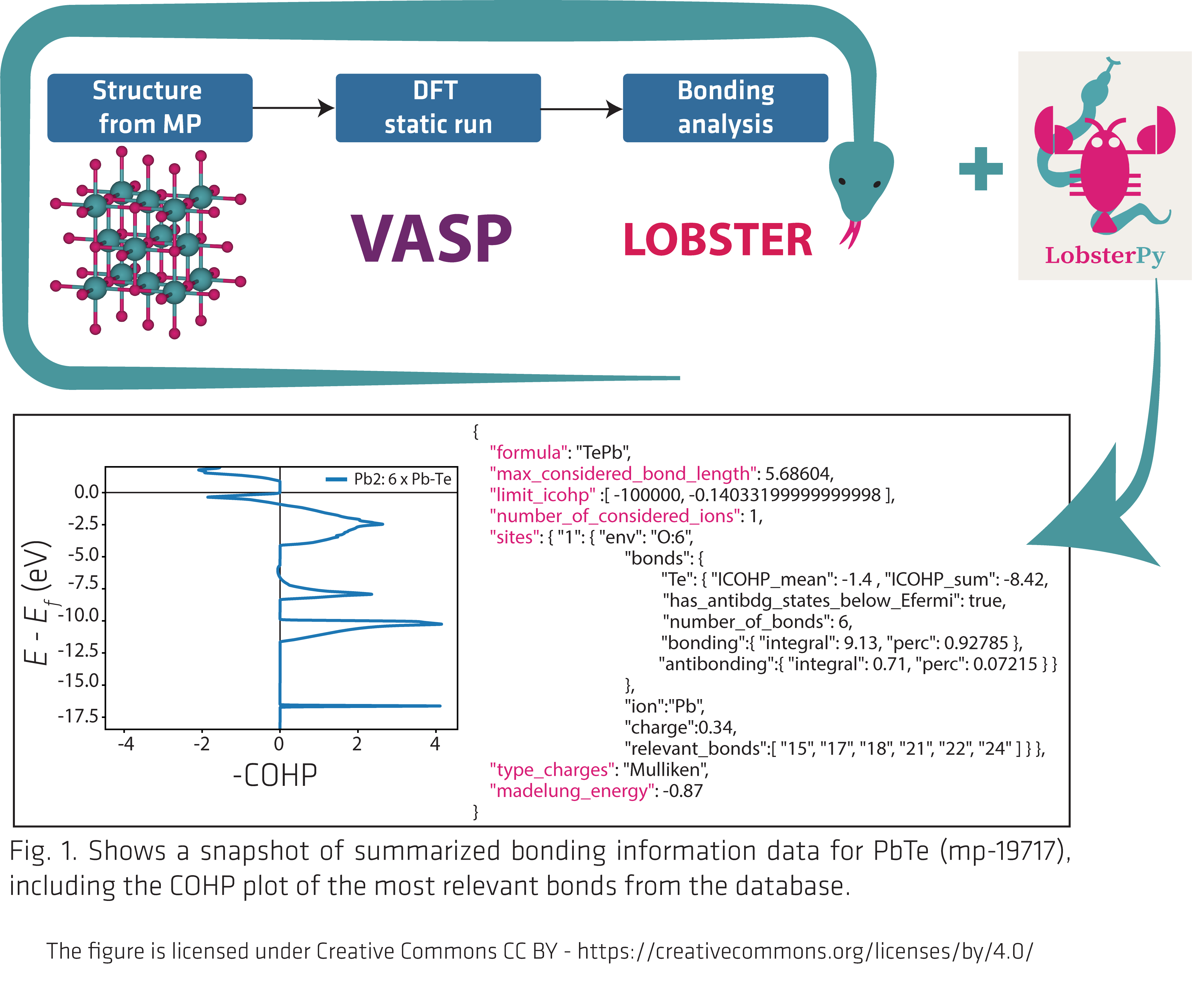
In this study, we harnessed the capabilities of the LOBSTER[1–4] software package, which takes modern density functional theory (DFT) data and transforms it into a form that reveals the bonding scenario in the materials. LOBSTER allows one to peer into the bonding world that holds the atoms together in solid-state materials by projecting plane wave-based wave functions onto a local, atomic orbital basis. The critical component necessary for this research is the development of a fully automatic workflow[5] that combines the VASP[6–8] and LOBSTER computations. Another essential component is the LobsterPy[9] package, which automates analyzing a vast number of output files to populate the dataset.
The final database is provided in two forms, both as JSON files. The first contains only summarized information necessary for gaining a quick insight into the chemical bonding scenario of the compound in question. This provides crucial information like, the number of ions, covalent bond strengths, coordination environment for ions, electrostatic charge of the structure, bonding and antibonding contributions to the bonds, and many more based on our quantum-chemical computations. The second database contains data from all important LOBSTER computations, including the calculation settings used, which are essential to reproduce our results. Please check our article and the codes provided for more details on where to find these data and how to access them.
Rigorous testing has ensured that the data we provide are reliable, and it also guarantees that the information contained within the database is of the highest quality, ready to drive further scientific discovery. We also demonstrate the dataset’s potential, where we leveraged the bonding descriptors (from our dataset) to construct a machine learning (ML) model for predicting phononic properties. The results showed a 27% increase in prediction accuracy compared to a benchmark model that did not rely on quantum-chemical bonding features.
Finally, let’s not forget about the critical component of the open-access code contributions that made our work reproducible. Getting them ready to be made openly accessible was filled with unseen hours of coding, extending existing code capabilities, code reviews, and rigorous testing. Thanks to this effort, we can now distribute the dataset and the codes to the data-driven materials research community. We believe that this will further advance the material informatics field.
We also plan to make our data accessible via a database server platform for seamless access. Stay tuned for further updates from us.
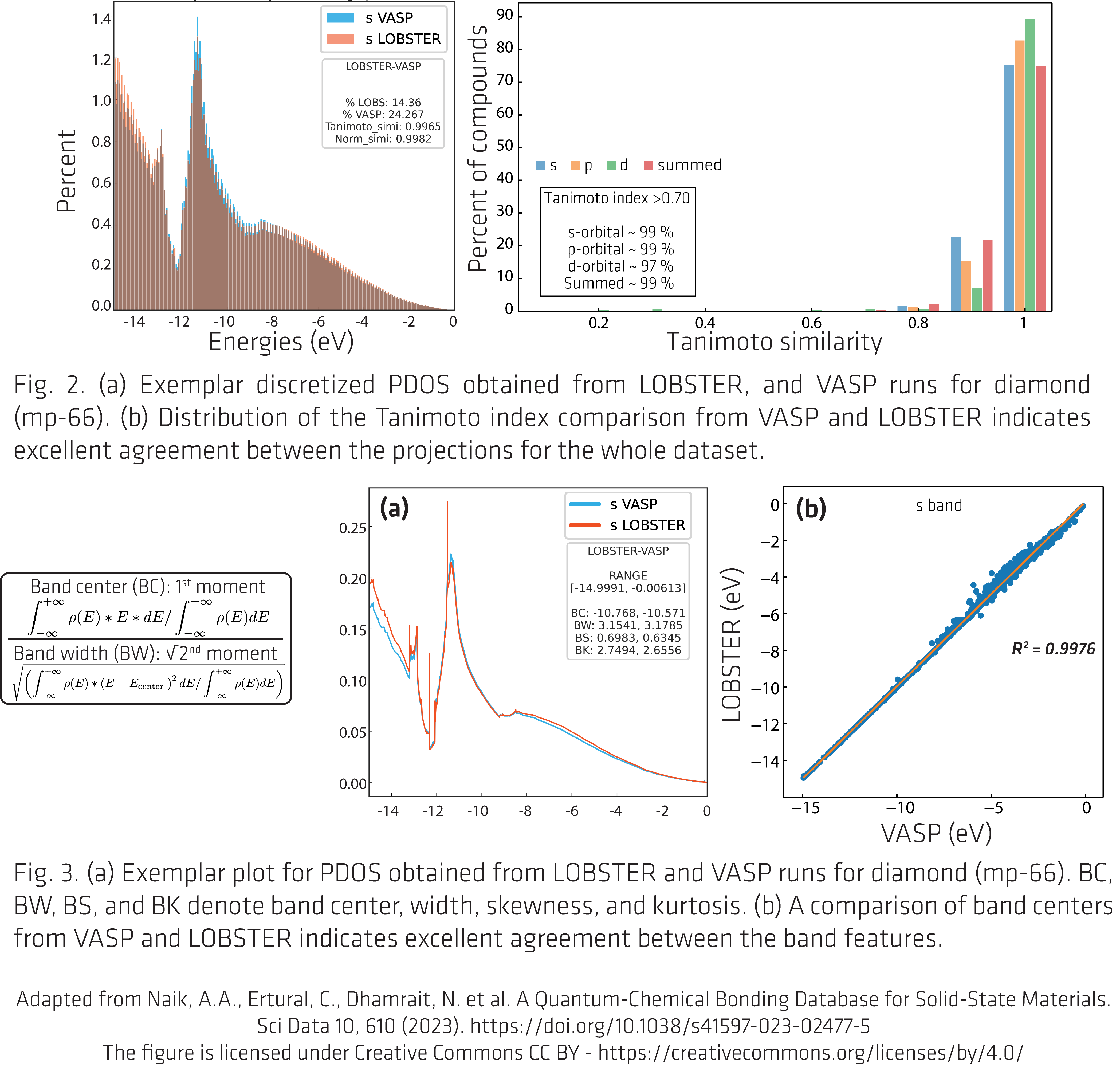
Figures 2 and 3 are adapted from our article. Link to our article: https://www.nature.com/articles/s41597-023-02477-5
All figures are licensed under Creative Commons CC BY https://creativecommons.org/licenses/by/4.0/.
References
[1] S. Maintz, V. L. Deringer, A. L. Tchougréeff, R. Dronskowski, J. Comput. Chem. 2016, 37, 1030–1035.
[3] V. L. Deringer, A. L. Tchougréeff, R. Dronskowski, J. Phys. Chem. A 2011, 115, 5461–5466.
[4] S. Maintz, V. L. Deringer, A. L. Tchougréeff, R. Dronskowski, J. Comput. Chem. 2013, 34, 2557–2567.
[6] G. Kresse, J. Furthmüller, Phys. Rev. B 1996, 54, 11169–11186.
[7] G. Kresse, J. Furthmüller, Comput. Mater. Sci. 1996, 6, 15–50.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Genomics in freshwater and marine science
Publishing Model: Open Access
Deadline: Jul 23, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in