Using a novel artificial protein scaffold to streamline metabolic flux: Accurate assembly of intracellular metabolic key nodes
Published in Bioengineering & Biotechnology

If a cell is a micro-factory, the enzymes in the cell are the machines. These machines catalyze numerous reactions inside the cells, many of which are tightly regulated[1]. Membrane-containing or membrane-free organelles, metabolons, multidomain synthases, or multienzyme complexes are formed through self-assembly to achieve a local confinement of the enzymatic activity, as one of the ways of regulating the chemical reactions[2-4]. Enzyme assembly also brings about higher catalytic efficiency. However, this level of regulation is often lost, or neglected, when enzymes are cloned and expressed in a heterologous host cell or in a test tube[5].
In this work, we adopted a novel protein-protein interaction based on a pair of simple peptide tags RIAD and RIDD, which enables the assembly of multiple enzymes in a sequential pathway. RIDD is a peptide consisting of forty-four amino acids derived from cAMP-dependent protein kinase A; RIAD is an amphiphilic peptide consisting of only 18 amino acids derived from a kinase-anchored protein. RIDD spontaneously forms a stable dimer in physiological conditions, subsequently, RIAD binds to the RIDD dimer to form a stable trimerization structure. Different expression ratios of multiple enzymes can be assembled by fusing the pair of peptide tags. We observed that RIAD and RIDD can effectively mediate the formation of enzymes with various structures in vitro. By adjusting the ratio of catalytic subunits, the efficiency of multi-enzyme catalytic reaction can be artificially regulated, and higher degree of enzyme assembly correlates with more efficient intermediate transfer and higher overall catalytic efficiency of the cascade reactions.
There is a practical need for enzyme assembly. Terpenoids are the most abundant class of compounds in nature and are widely used in biomedicine, food, et al. The biosynthesis of terpenoids can be divided into upstream and downstream, the upstream provides the precursors IPP and DMAPP, the downstream produces a variety of terpenoids by modification of the precursors IPP and DMAPP. Idi is the last enzyme of the upstream, which is free in the cytoplasm; CrtE is the first enzyme in the downstream of carotenoid biosynthesis, however, the enzyme is distributed on the cell membrane. A critical question however resides when all the enzymes with highest catalytic efficiency in the terpenoid pathway are expressed in a bacterial host, for example, E. coli: the accumulation of IPP and DMAPP inhibits cell growth, due to insufficient coupling of the upstream and downstream pathways.
The RIAD-RIDD assembly system perfectly fits here. By constructing Idi-CrtE multi-enzyme complex using the RIAD-RIDD pair, we found many Idi protein relocated to the cell membrane, and the synthesis flux of carotenoids greatly improved. The Idi-CrtE multi-enzyme complex links the upstream and downstream physically, and the accurate assembly of intracellular metabolic key nodes streamline the target metabolic fluxes. Subsequently the astaxanthin production increased by 2.7 times and the carotenoid production increased by 5.7 times. In a lycopene high-yield strain of Saccharomyces cerevisiae, lycopene production increased by 58% to 2.3 g/L, which is the highest yield of lycopene produced by S. cerevisiae.
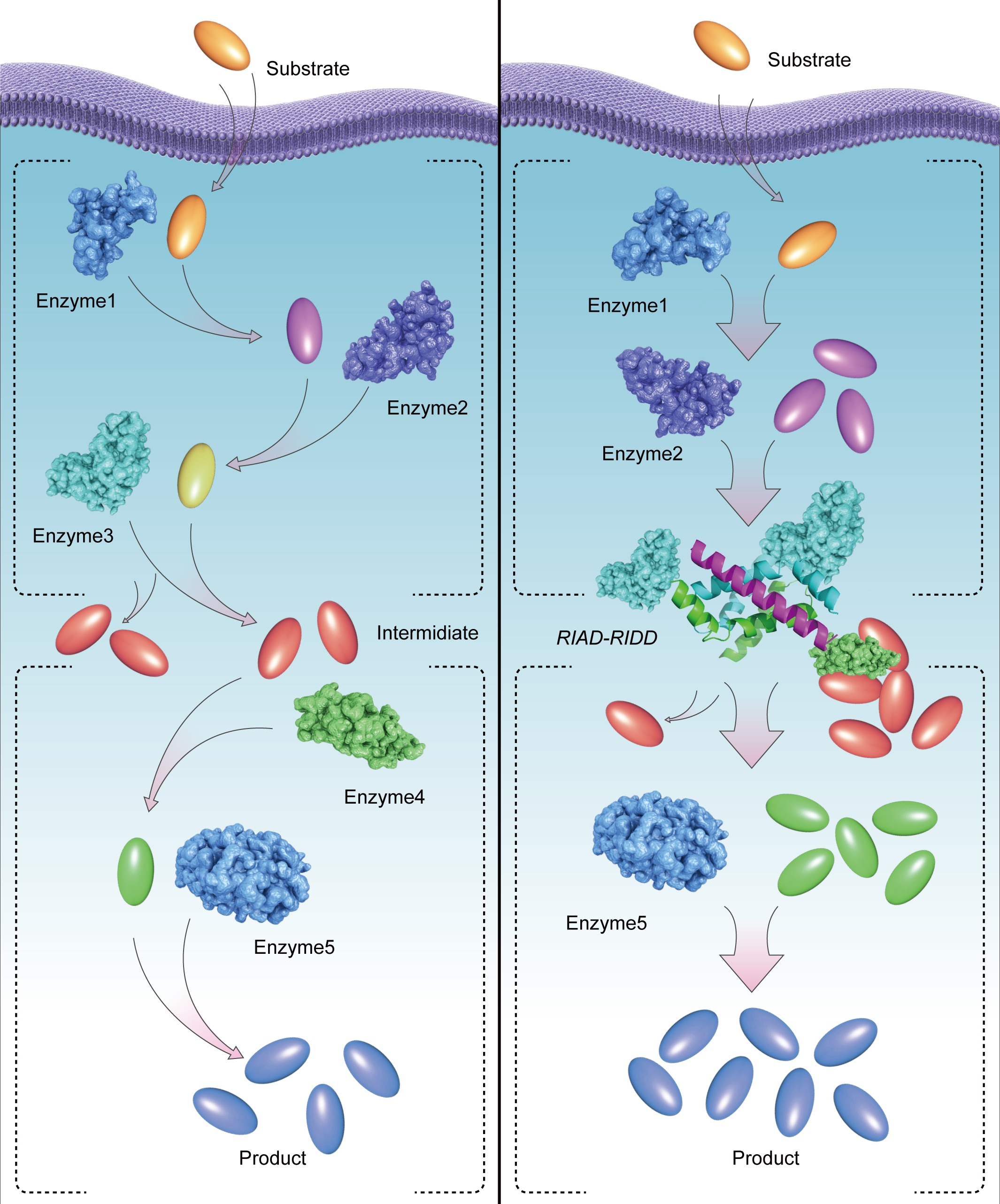
Our work showcases the importance of the location of an enzyme to its role in the sequential enzyme reaction. Further, by constructing a multi-enzyme complex to link the key metabolic nodes of biosynthesis to increase the production of target pathway, especially for artificial metabolic systems with different spatial distribution enzymes, our method not only solved the problem of substrate transportation, but also reduced the imbalance of artificial metabolic system. Synthetic multienzyme complexes, mimicking metabolons and natural multienzyme complexes, hold promise in optimizing the flux of natural product biosynthesis.
Written by Tian Ma, Jiang Xia, and Tiangang Liu.
Link to our paper: https://www.nature.com/articles/s41467-019-12247-w
Reference
1. Ajikumar, P.K. et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330, 70-74 (2010).
2. Dutta, S. et al. Structure of a modular polyketide synthase. Nature 510, 512-517 (2014).
3. Lomakin, I.B., Xiong, Y. & Steitz, T.A. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 129, 319-332 (2007).
4. Polka, J.K., Hays, S.G. & Silver, P.A. Building spatial synthetic biology with compartments, scaffolds, and communities. CSH Perspect. Biol. 8, a024018 (2016).
5. Dahl, R.H. et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039-1046 (2013).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in