Why is Singapore Identified in Global Research as Number One? How Physical Activity and Education Excellence Created a Global Leader

Colorectal cancer is the third-leading cause of cancer-related deaths in the United States. Like many other cancers, whether the cancer has spread away from the primary site to regional lymph nodes (i.e. lymph node metastasis, LNM) is a key factor in staging, treatment decision making and prognosis. Detecting LNM is an involved process dependent on adequate sampling by the surgeon, and a thorough review by the pathologist. Unfortunately, this is a challenging process with several opportunities for missed diagnoses. For example, in order to identify that cancer has spread to a lymph node, the affected node must first be excised, a process subject to sampling bias as not every regional lymph node will be removed at the time of surgery. Then, even for lymph nodes that are removed, the node must be appropriately identified and isolated during processing of the surgical specimen, sectioned (cut) in a location that has tumor cells, and a pathologist must identify those tumor cells on review of that section under the microscope. Importantly, even patients who have no identified spread to lymph nodes still have 5-year mortality rates of 20-30%, which is thought to be largely due to missed identification of lymph node involvement.1,2
Our motivation for this project was to explore whether the presence of LNM could be further informed by using a deep learning-based model examining the primary tumor (i.e., the site of the original tumor with no analysis of lymph node tissue itself). Such a prediction tool could help augment identification of lymph node metastasis, perhaps by flagging cases at higher risk of LNM for more careful review or sampling. While previous research has demonstrated some associations of established histologic features with LNM, no system currently exists in practice to utilize these features in a systematic fashion to generate an accurate risk assessment for LNM.
Deep learning has shown great promise in many areas of medical imaging analysis, including in prediction tasks involving computational pathology, but to this point has struggled to show improved predictive power over models only using known clinicopathologic associations.3,4 Deep learning models are powerful learners of patterns in data, and a deep learning model trained directly to produce a risk score for some outcome of interest may simply relearn the known associations with that outcome of interest. Therefore, it is unsurprising that deep learning-based risk scores may fail to boost predictive power when added to these other known associations.
In “Predicting lymph node metastasis from primary tumor histology and clinicopathologic factors in colorectal cancer using deep learning”, published in Nature Communications Medicine, we develop a system to control for baseline variables while selecting informative deep learning features, thus forcing the deep learning features to represent concepts that are independent of the known variables. In this research effort involving Google Health, Verily, Stanford University School of Medicine, and Medical University of Graz, we extract thousands of small image crops, or patches, from tumor-containing regions of colorectal tissue. Each of these patches is passed through a previously trained deep learning model to obtain an “embedding” for each patch, representing the visual features within that patch. We then perform clustering (via k-means) on these patch embeddings to assign each patch to one of 200 clusters. Previous work has shown that clustering embeddings in this way results in clusters that contain visually-similar features, enabling further interpretability and the possibility of discovering and describing new morphological biomarkers.5 For each cancer case, we then record the fraction of patches that belong to each cluster, which we term the “cluster quantification” of that case.
To build our final predictive model, we select the single machine-learned feature (based on cluster quantification fraction for that feature) that achieves best predictive performance when combined with all known baseline variables available. By incorporating the baseline variables in the multivariable logistic regression at the time of feature selection, we are “controlling” for these features in order to select the machine-learned feature that provides the most independent signal in addition to the known variables. We then repeat this process 4 more times, with each successive step including the previously selected machine-learned features along with the baseline features. By the end of the process, we have selected 5 machine-learned features that provide the largest boost in predictive power when combined with the baseline variables, minimizing the possibility of selecting features directly related to the baseline variables themselves.
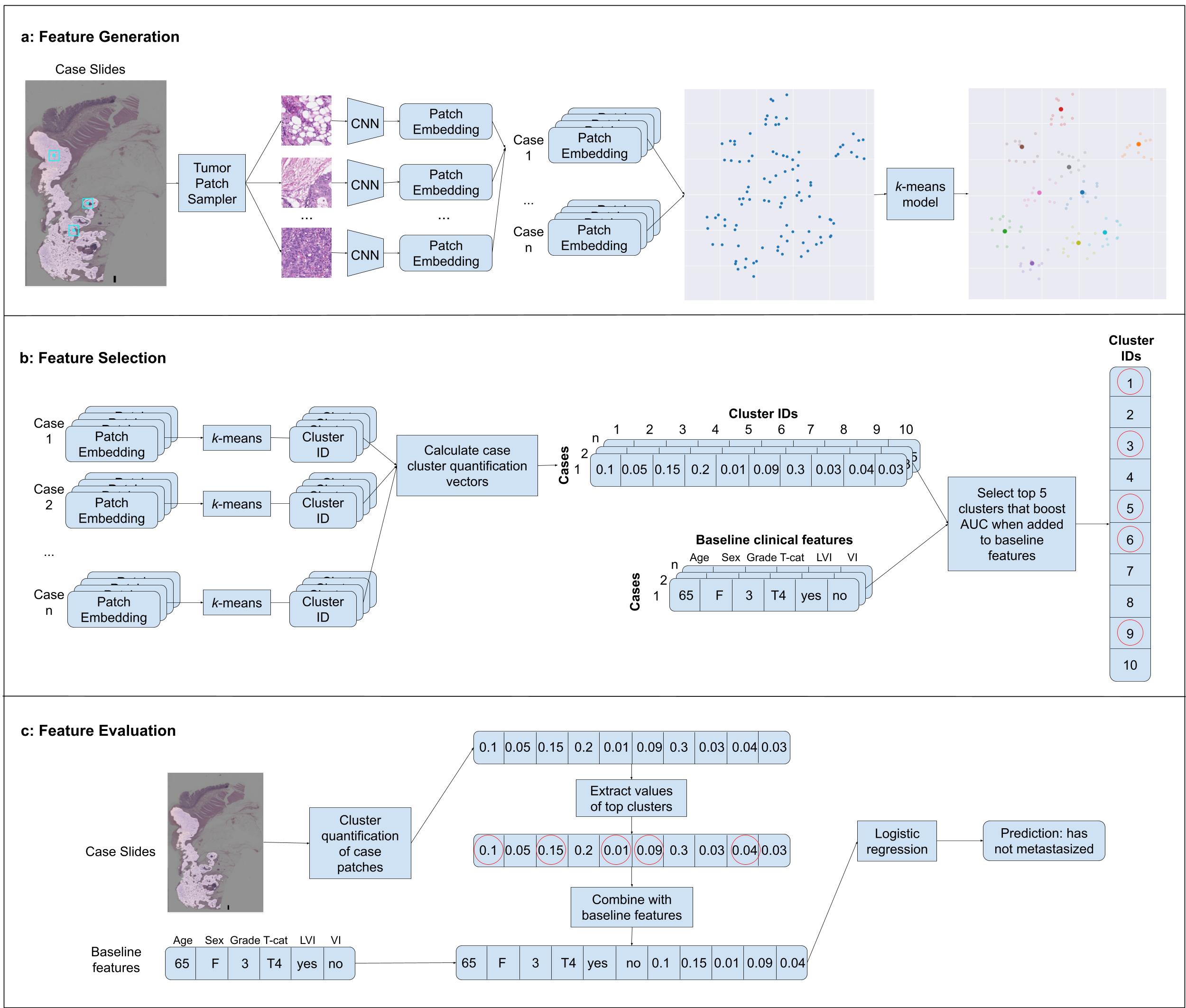 Overview of the development of our machine learning model. Panel a shows machine-learned feature generation in which patches from tumor-containing regions of colorectal cancer cases are passed through a deep learning model to obtain an embedding vector for each patch. These embeddings are then clustered using a k-means algorithm. Panel b shows the selection of machine-learned features wherein a cluster quantification vector is computed for each clinical case, and the top 5 clusters are sequentially chosen that maximize predictive performance when combined with baseline clinical features in a logistic regression model. Panel c demonstrates final model prediction whereby cluster quantification values for the selected clusters are added to the baseline clinical features, and this combined feature vector is fed through a logistic regression model to obtain a prediction.
Overview of the development of our machine learning model. Panel a shows machine-learned feature generation in which patches from tumor-containing regions of colorectal cancer cases are passed through a deep learning model to obtain an embedding vector for each patch. These embeddings are then clustered using a k-means algorithm. Panel b shows the selection of machine-learned features wherein a cluster quantification vector is computed for each clinical case, and the top 5 clusters are sequentially chosen that maximize predictive performance when combined with baseline clinical features in a logistic regression model. Panel c demonstrates final model prediction whereby cluster quantification values for the selected clusters are added to the baseline clinical features, and this combined feature vector is fed through a logistic regression model to obtain a prediction.
We found on both internal and external validation sets that our machine-learned features provided signal independent of LNM and boosted predictive performance for LNM when added to baseline variables. It is important to note that the predictive power remained modest (AUROC for the combined model was 0.740 on our primary external validation set) and that this model is clearly not a substitute for careful surgical excision and pathologic examination of nodes. Still, this type of information could potentially provide useful signal for triaging and further follow up. We also found that our model’s output could risk stratify stage II (no detected LNM) and stage III (detected LNM) cases in our internal dataset, suggesting the potential for such a model to provide important prognostication and potentially aid in risk-based clinical decision making.
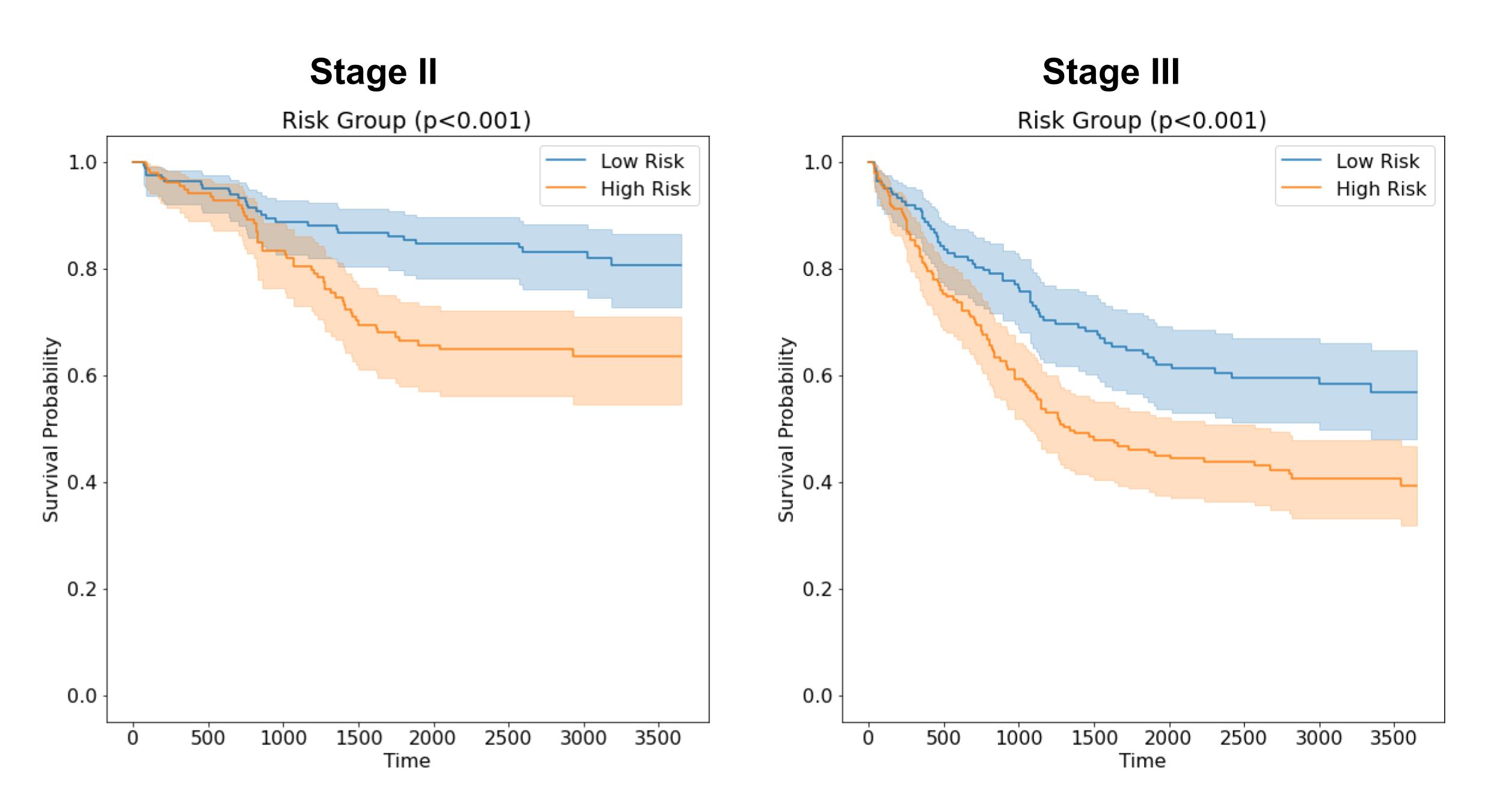
Kaplan-Meier survival curves for stage II and stage III colorectal cancer cases, grouped into high and low risk by machine learning model risk score.
Additionally, since the clustering of embeddings produces visually similar features, it is possible to visually analyze the 5 selected machine-learned clusters. Interestingly, one of these clusters appears to share similarities with recently described features identified independently and found to be associated with metastasis and adverse outcomes in colorectal cancer.5,6,7,8
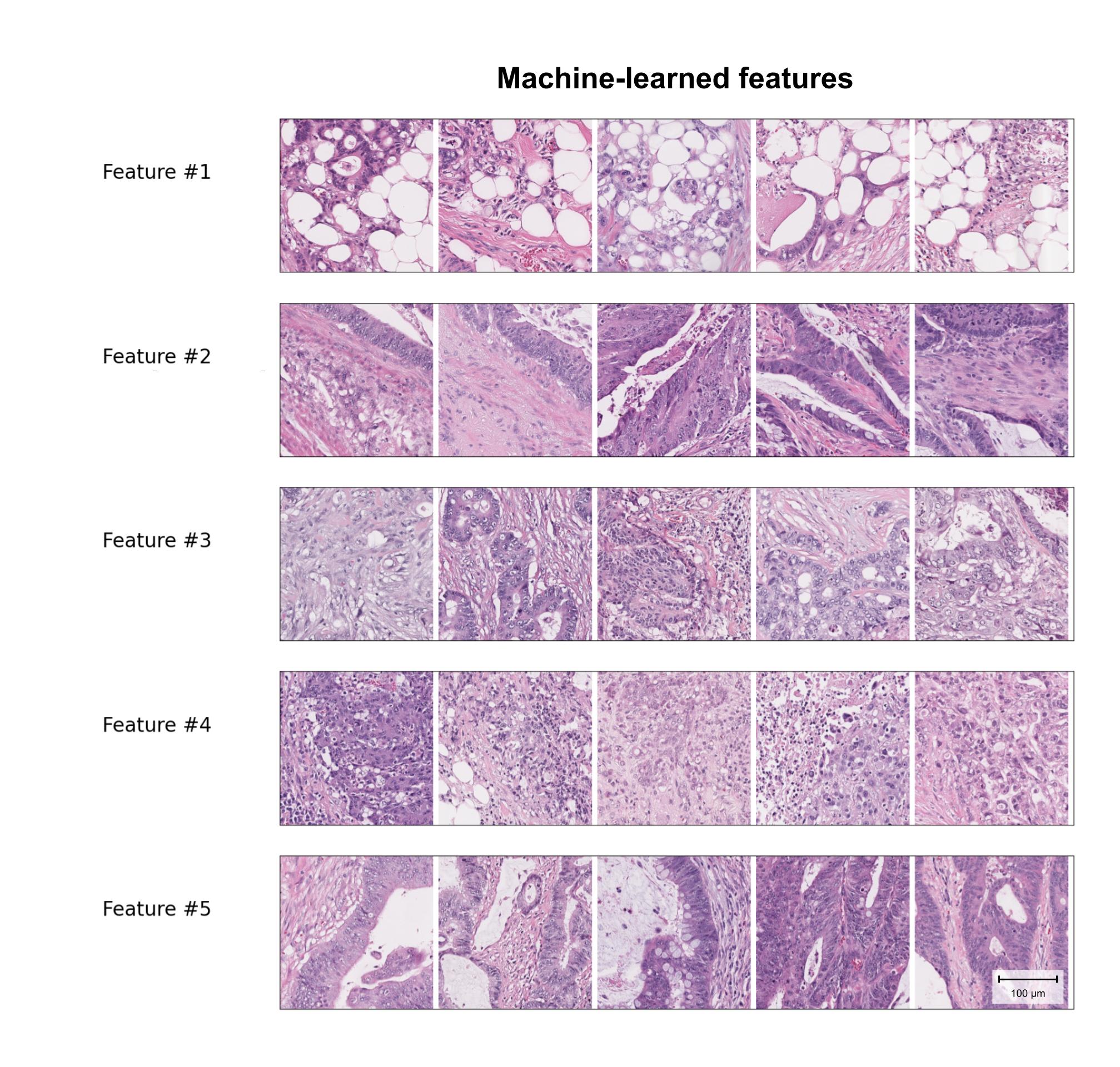
Sample patches from the top 5 selected machine-learned features. Feature #1 shares some characteristics with a previously described Tumor Adipose Feature, recently identified as a potential risk factor for poor outcomes in colorectal cancer.
In this study we identified 5 machine-learned histopathologic features that add predictive performance to known clinicopathologic features in the prediction of LNM. While the predictive boost is modest, the findings indicate presence of additional signal for LNM prediction beyond currently appreciated features, representing an opportunity for future research into morphological associations with pathological processes such as lymph node metastasis. With further refinement and validation, a predictive model such as this could also prove useful for informing clinical decision making, such as identifying high risk patients with Stage II cancer who may benefit from adjuvant chemotherapy, or flagging patients with T1 cancer undergoing endoscopic resection who may benefit from surgical resection with full lymph node staging. More generally, the findings are also consistent with the intriguing possibility that tumors acquire properties and morphological characteristics associated with metastatic potential, perhaps even prior to metastasis itself - such characteristics will be interesting to explore across additional cancer type as well.

A selective open access journal from Nature Portfolio publishing high-quality research, reviews and commentary across all clinical, translational, and public health research fields.
With Collections, you can get published faster and increase your visibility.
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Publishing Model: Open Access
Deadline: Jun 01, 2026

We use cookies to ensure the functionality of our website, to personalize content and advertising, to provide social media features, and to analyze our traffic. If you allow us to do so, we also inform our social media, advertising and analysis partners about your use of our website. You can decide for yourself which categories you want to deny or allow. Please note that based on your settings not all functionalities of the site are available.
Further information can be found in our privacy policy.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in