Widespread HGT in insects: a foreign gene enhances male courtship in lepidopterans
Published in Ecology & Evolution
Over the years, the evolutionary significance of horizontal gene transfer (HGT) in bacterial genome evolution is well-established, and several recent studies suggest that HGT may also play an important role in the evolution of eukaryotic genomes (e.g., Husnik and McCutcheon, 2018; Perreau and Moran, 2022). For example, carotenoid biosynthesis genes transferred from fungi to aphids contribute to aphid body coloration (Moran and Jarvik, 2010). However, a systematic investigation of HGT-acquired genes in insect genomes, including an examination of their fates and the functions that these foreign genes have contributed to, is still lacking.
In the summer of 2020, Shenlab at Zhejiang University started this project in collaboration with Rokaslab at Vanderbilt University, for which we took advantage of 218 high-quality genomes of insects representing 11 out of all 16 species-rich insect orders (i.e., orders with >1,000 described species) (Stork, 2018), to systematically identify HGT-acquired genes, and study their evolution and function. At the beginning of 2021, we identified a list of over 1,000 HGT-acquired genes that were likely acquired from non-metazoan sources. In the middle of 2021, another two studies found that transferred genes can have ecologically diverse functions including detoxification (Xia et al., 2021) and defense (Gasmi et al., 2021). Given these three important HGT cases reported in previous studies, we were excited to check whether our list included these three cases or not. Surprisingly, we found that the list of HGT-acquired genes not only included three examples cited in the previous sentences but also provided many additional diverse functions. Then, we naturally asked three questions: (i) What is the distribution of horizontally acquired genes across major insect groups? (ii) What factors contribute to the adaptation of HGTs in insect genomes? (iii) What are the biological functions of HGTs in insects?
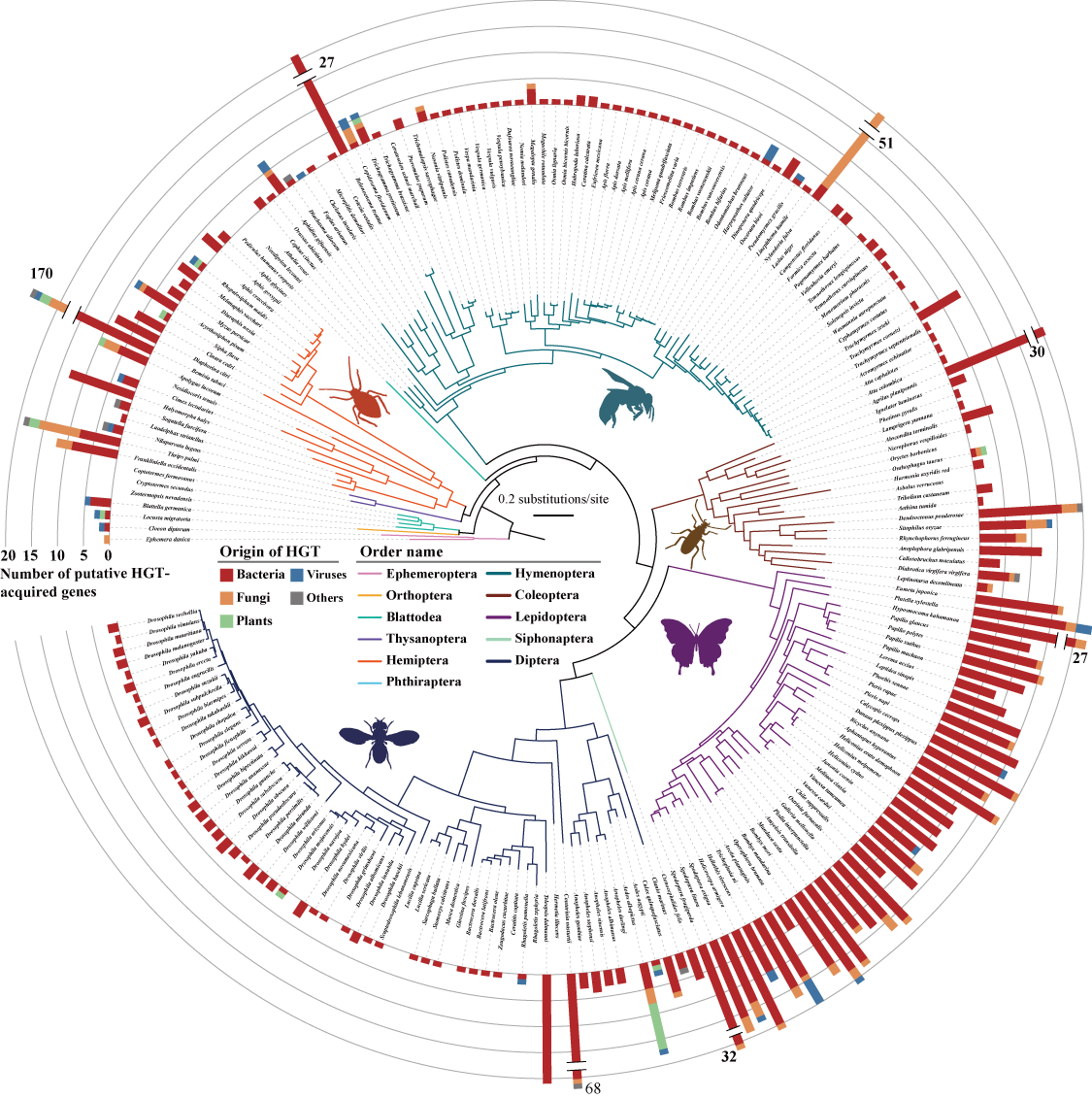
First, we found a total of 1,410 genes in 192 insect genomes that were likely acquired via 741 distinct events from non-metazoan sources, including 1,115 (79.0%) genes from Bacteria, 194 (13.8%) genes from Fungi, 43 (3.0%) genes from plants, 36 (2.6%) genes from viruses, and 22 (1.6%) genes from other lineages. In addition, we found that the order Lepidoptera acquired by far the highest average number of HGT-acquired genes (16 genes per species), followed by the orders Hemiptera (13 genes per species), Coleoptera (6 genes per species), Hymenoptera (3 genes per species), and Diptera (2 genes per species).
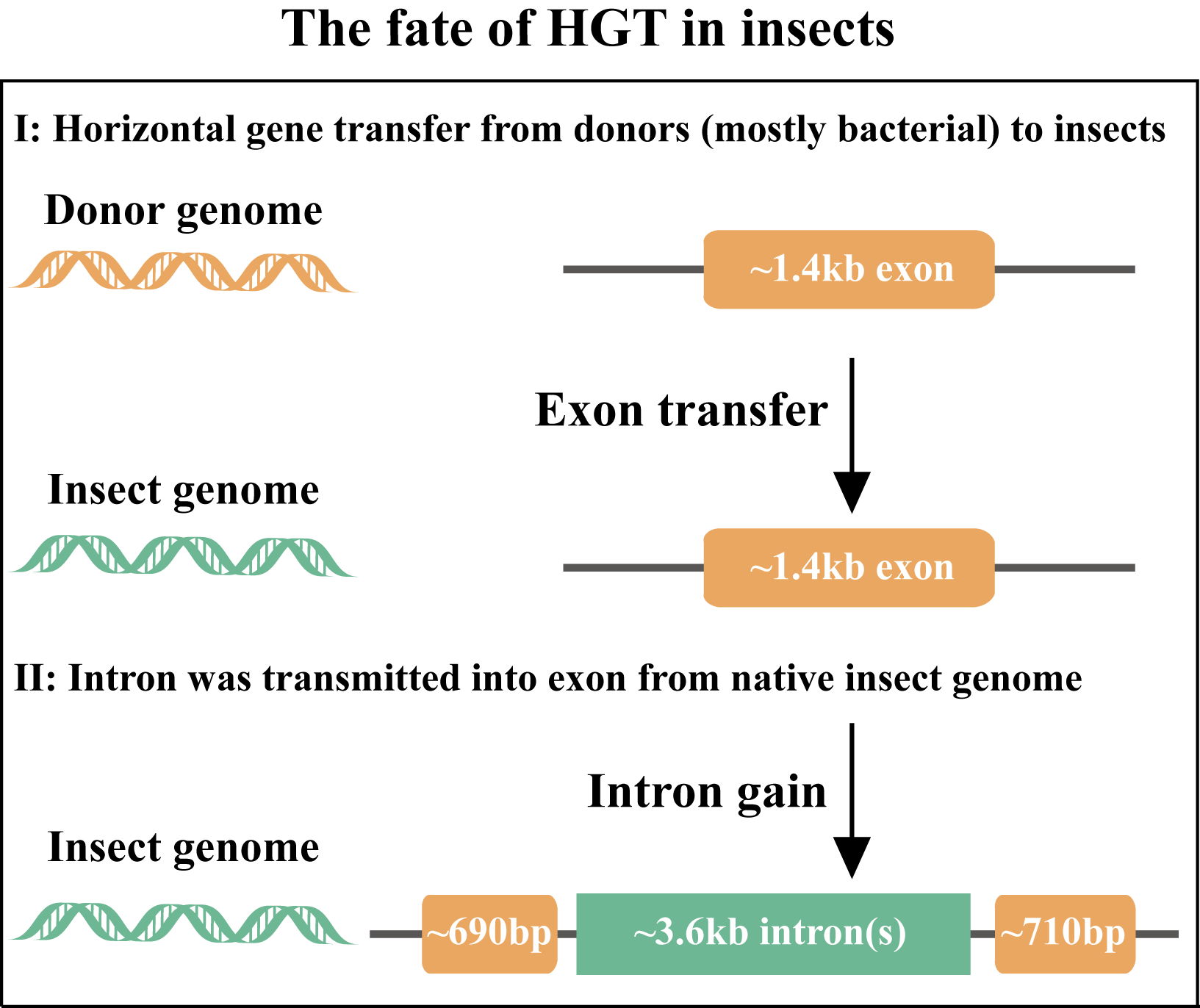
Second, by characterizing the gene structures of 1,410 HGT-acquired genes in the putative donor and recipient genomes as well as of all native insect genes, we found that 849 / 1,410 HGT-acquired genes contain repeat-rich introns, which were likely acquired from the native insect genomes after the initial gene transfers. Moreover, a comparison of gene structures between HGT-acquired genes in the context of divergence times shows that intron gain events occurred over evolutionary time, which enabled these foreign genes to increase their lengths toward the average length of native genes. More importantly, HGT-acquired genes containing introns exhibited substantially higher expression levels than genes lacking introns, which is consistent with previous studies in diverse organisms (green microalgae, plants, and insects) showing that intron gains can enhance gene expression levels (Baier et al., 2018; Husnik et al., 2013; Rose et al., 2011). These results suggest that intron gains were likely involved in HGT adaptation.
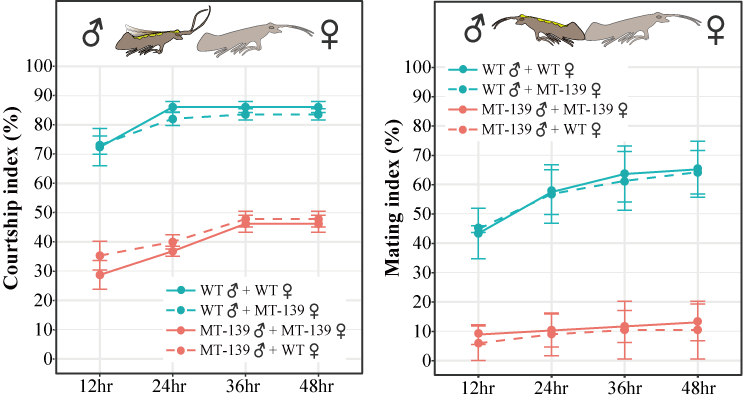
Third, the set of 1,410 HGT-acquired genes not only included the majority (~85%) of previously reported cases, but also provided additional diverse functions, including but not limited to metabolism, courtship behavior, nutrition, adaptation to an extreme environment, growth, and development. Among 1,410 HGT-acquired genes, the most widely distributed HGT-acquired gene in insect genomes revealed by our analyses was LOC105383139, an uncharacterized gene acquired from a Listeria-related bacterial donor in the ancestor of butterflies and moths. We took this gene to Dr. Jianhua Huang at Zhejiang University and asked whether his group could use the CRISPR-Cas9 system to dissect the function of this gene in the non-model species diamondback moth Plutella xylostella, a serious agricultural pest of crucifer vegetables. Surprisingly, on the day before Christmas in 2021, Jianhua told us that male diamondback moths lacking LOC105383139 reduced male courtship and mating behaviors, but have no significant differences in developmental phenotypes, including body size, feeding, and movement. We also found that the role of the gene LOC105383139 in two closely related butterflies (Heliconius melpomene and Heliconius cydno) is consistent with our finding that this foreign gene can enhance male courtship behavior in diamondback moths, using the publicly available courtship data and transcriptome data (eye and brain) in butterfly courtship situations.
Collectively, we provided a comprehensive resource of HGT events and their fates in insects, coupled with our discovery of a functional role for a horizontally acquired gene (male courtship behavior) in a major insect lineage (moths and butterflies).
The paper published in Cell can be found here.
References
Baier, T., Wichmann, J., Kruse, O., and Lauersen, K.J. (2018). Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 46, 6909–6919.
Gasmi, L., Sieminska, E., Okuno, S., Ohta, R., Coutu, C., Vatanparast, M., Harris, S., Baldwin, D., Hegedus, D.D., Theilmann, D.A., et al. (2021). Horizontally transmitted parasitoid killing factor shapes insect defense to parasitoids. Science (80-. ). 373, 535–541.
Husnik, F., and McCutcheon, J.P. (2018). Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 16, 67–79.
Husnik, F., Nikoh, N., Koga, R., Ross, L., Duncan, R.P., Fujie, M., Tanaka, M., Satoh, N., Bachtrog, D., Wilson, A.C.C., et al. (2013). Horizontal Gene Transfer from Diverse Bacteria to an Insect Genome Enables a Tripartite Nested Mealybug Symbiosis. Cell 153, 1567–1578.
Moran, N.A., and Jarvik, T. (2010). Lateral Transfer of Genes from Fungi Underlies Carotenoid Production in Aphids. Science (80-. ). 328, 624–627.
Perreau, J., and Moran, N.A. (2022). Genetic innovations in animal–microbe symbioses. Nat. Rev. Genet. 23, 23–39.
Rose, A.B., Emami, S., Bradnam, K., and Korf, I. (2011). Evidence for a DNA-Based Mechanism of Intron-Mediated Enhancement. Front. Plant Sci. 2.
Stork, N.E. (2018). How Many Species of Insects and Other Terrestrial Arthropods Are There on Earth? Annu. Rev. Entomol. 63, 31–45.
Xia, J., Guo, Z., Yang, Z., Han, H., Wang, S., Xu, H., Yang, X., Yang, F., Wu, Q., Xie, W., et al. (2021). Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184, 1693-1705.e17.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in