World Cancer Day: The expression of cuproplasia-related genes can predict the patient outcome in pan-cancer
Published in Cancer
Cuproplasia-related genes: Why are they important to identify cancer patients who may have a better outcome?
Cuproplasia is copper-dependent cell-growth and proliferation in cancer, and copper is a redox cofactor for several copper-dependent enzymes and proteins, such as mitochondrial Cu transporter SLC25A3 and cytochrome c oxidase COX. Copper plays a critical role in angiogenesis, by influencing vascular endothelial growth factor (VEGF) protein and its mRNA expression. All these findings indicate the importance of copper as a factor for enzymes in mediating many vital cellular functions. However, the dysregulation of copper stimulates receptor tyrosine kinase (RTK) signalling pathways, inhibits key enzymes in cellular metabolism and respiration, and leads to oxidative stress and cytotoxicity. Some proteins maintain appropriate intracellular copper bioavailability in cells and ensure the metalation of copper-dependent enzymes. Key regulators of mammalian copper homeostasis are blood carrier protein ceruloplasmin (CP), SLC31A1 (also known as CTR1, a principal copper uptake transporter), and exporting proteins ATP7A and ATP7B, which possess both copper export and metallochaperone functions. The expression dysregulation in those regulating proteins cause copper accumulation or deficiency pathologies, leading to diseases. Thus, proteins and genes related to copper and cuproplasia play a critical role in cancer initialisation and progression, and they can be used to predict the outcome of cancer patients.
How was the study conducted?
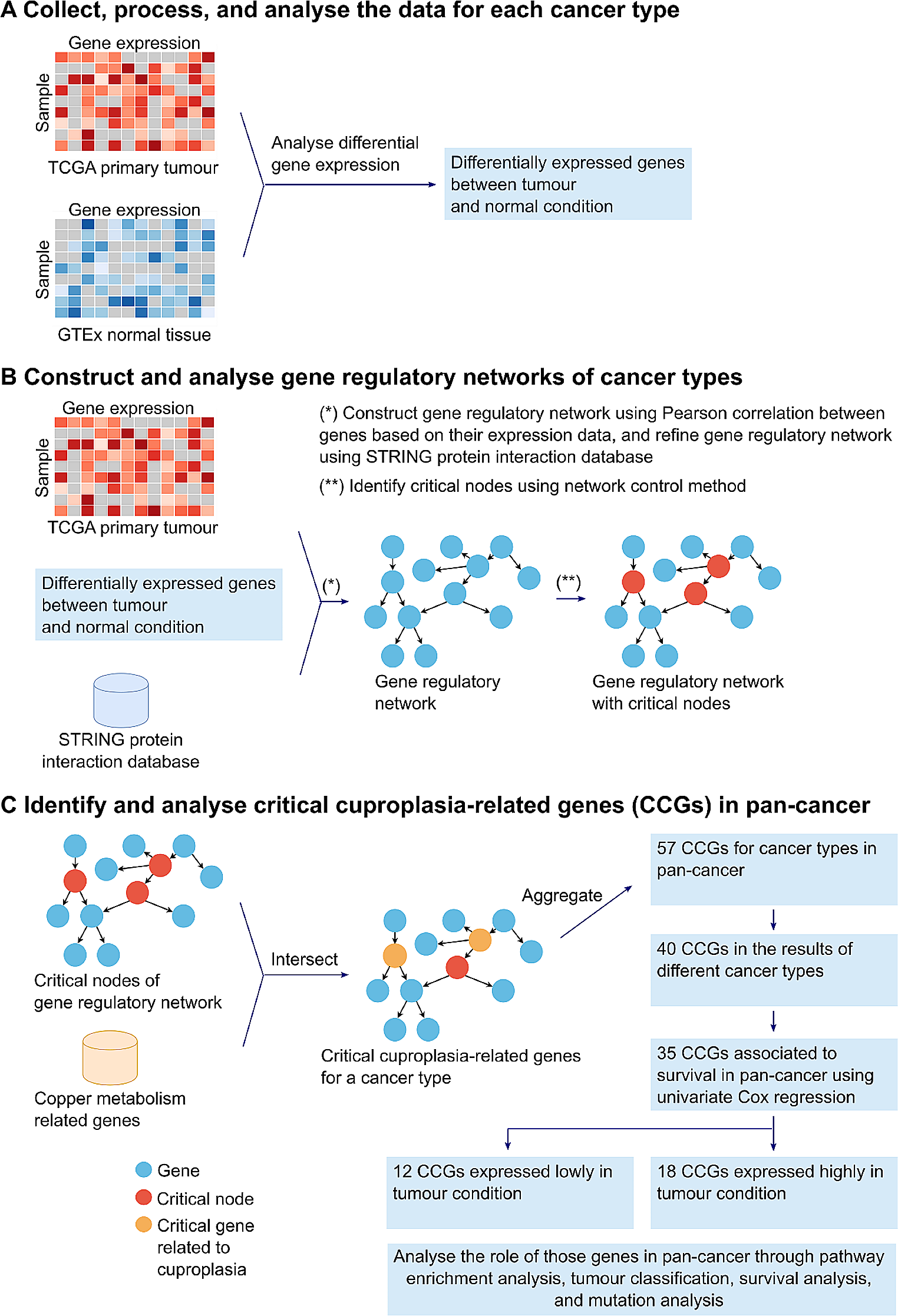
A comprehensive analysis was performed on cuproplasia-related genes in cancer to better understand their potential roles, effects, and associations with patient survival. Firstly, 23 gene regulatory networks were generated using the gene expression data of 23 tumours and normal tissue samples obtained from TCGA and GTEx respectively (Fig. 1A-B). To identify the critical cuproplasia-related genes (CCGs), we intersected the critical gene nodes of the estimated networks for each cancer type with the copper metabolism related genes from MSigDB. The critical gene nodes of each network were indicated by using the network control method. We then aggregated all the identified CCGs of the 23 cancer types into a single set and removed duplicated genes, resulting in 57 genes. Of these 57 genes, 40 genes occurred in multiple cancer types. We sought to further refine the list of CCGs to identify a potential specific subset of genes whose expression may be prognostic of survival in pan-cancer. Univariate Cox proportional hazards regression model based on the 40 genes revealed that 35 genes were significantly associated with survival (p < 0.05). Of these 35 genes, 18 genes were up-regulated and 12 down-regulated in tumours compared to matched normal tissue expression. Finally, the role of those genes in pan-cancer was explored through pathway enrichment analysis, tumour classification, survival analysis, and mutation analysis (Fig. 1C).
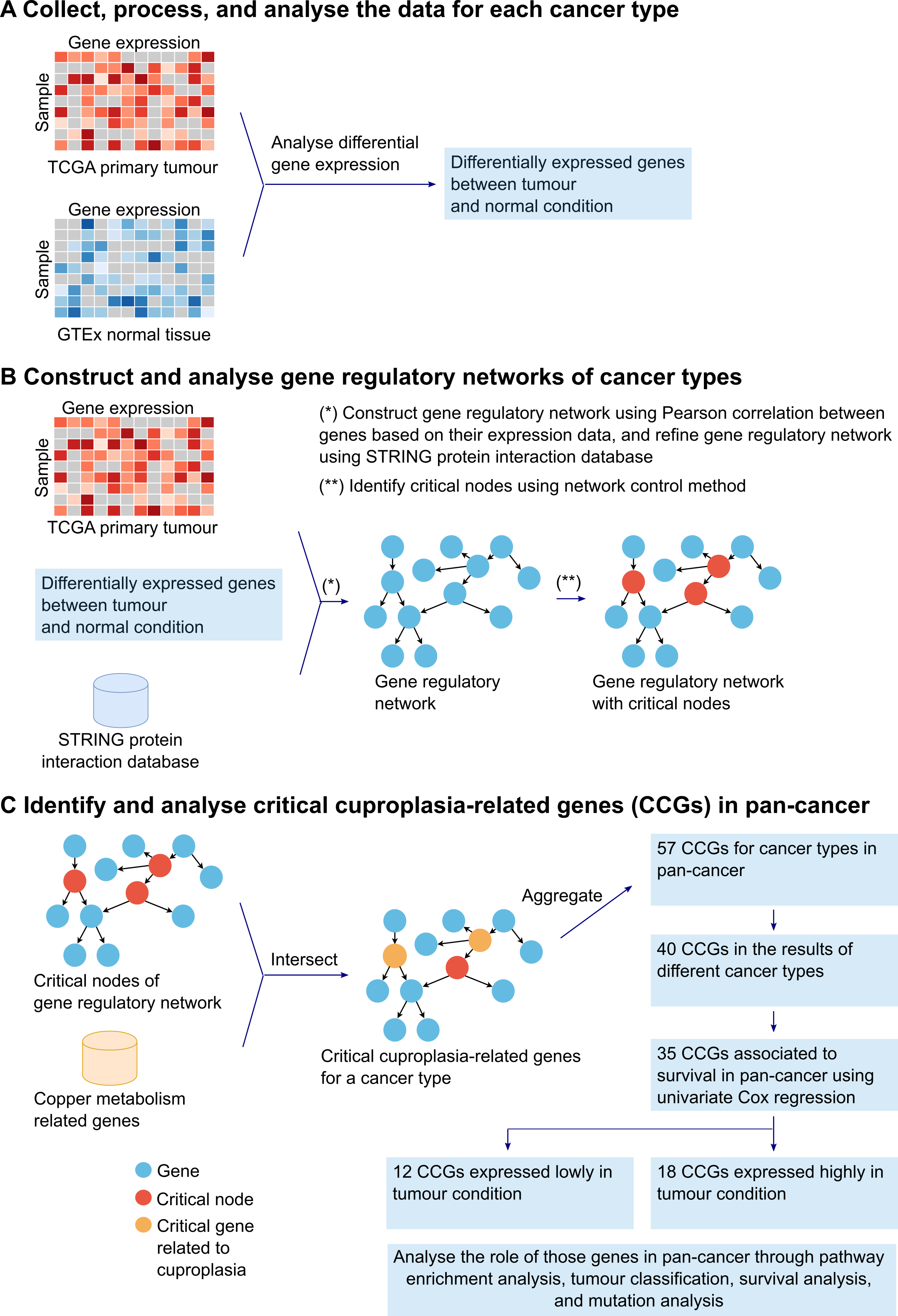
Fig. 1 Gene regulatory network methodology to identify 30 CCGs in pan-cancer
What did the study find?
From the 18 up-regulated and 12 down-regulated genes, we identified 8 gene pan-cancer specific signature that significantly improved survival (p = 1.88e-06; Fig. 2A). Patients whose tumour had high expression of MAP1LC3A, SNCA, MAPT and low expression of CDK1, AP1S1, CASP3, TMPRSS6, GSK3B inferred better overall survival than all other patients (Fig. 2A). Conversely patients who had low expression of MAP1LC3A, SNCA, MAPT and high expression of CDK1, AP1S1, CASP3, TMPRSS6, GSK3B, had the poorest overall survival. Further separating the genes into the 5 highly expressed and 3 lowly expressed genes also showed a significant improvement in survival (p = 1.91e-13 and p = 8.31e-10, respectively), however, resulted in poorer survival probability than the combined 8 gene signature (Fig. 2A-C). Based on these findings it is the combination of the 3 down-regulated and 5 up-regulated genes together that make up this novel signature that is more predictive of survival. Thus, our novel 8 CCGs could potentially be used to stratify patients prognostically regardless of cancer type. Interestingly, among these 8 CCGs MAP1LC3A, SNCA, CDK1, and CASP3 are involved in autophagic processes linked to cancer formation and progression. Importantly, SNCA has also been shown to play a potential role in pathological processes in distinct tumour types, including lung adenocarcinoma, ovarian cancer, and breast cancer. CDK1 dysregulation (i.e., a hallmark of cell cycle dysregulation) and GSK3B dysregulation observed in homeostatic dysfunction constitute key hallmarks of malignancies. Additionally, we previously demonstrated that copper chelation therapy inhibited GSK3B phosphorylation and in turn induced downregulation of PD-L1 and increased anti-cancer immune response. Regarding immune processes, high expression of AP1S1 in tumours was negatively correlated with immune infiltrating cells, and low expression of SNCA was correlated to low levels of immune infiltration, indicating that these genes play a key role in the anti-cancer immune response and immune evasion mechanism. Given the important roles of these genes in tumour formation and development, the use of therapeutic strategies targeting copper could have a strong effect on these genes and improve the prognosis of patients affected by different types of tumours.
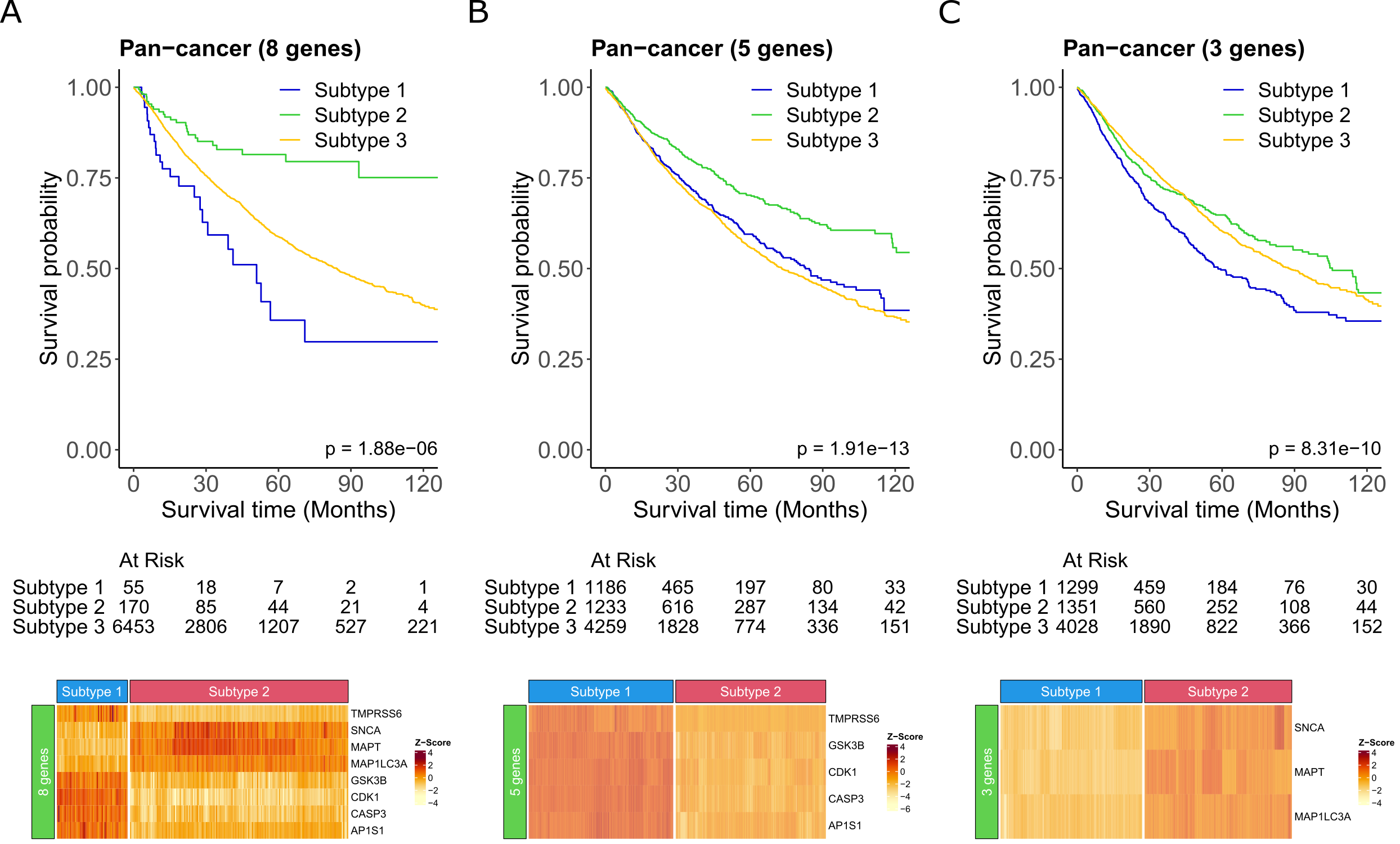
Fig. 2 A novel cuproplasia-related 8-gene signature is a potential biomarker of improved survival
What do these results mean?
We conducted for the first time a comprehensive pan-cancer analysis for genes involved in cuproplasia using a novel framework. An 8-gene signature related to cuproplasia was identified as a potential biomarker for predicting cancer patients’ survival. These findings provide novel insights into the effects of genes involved in cuproplasia on the molecular regulatory mechanisms of cancer development. Furthermore, the identified cuproplasia-related genes can be used by clinicians to understand which patients can benefit from using copper chelation therapy and identify novel combinatorial therapeutic strategies.
Follow the Topic
-
Human Genetics

This is a leading international journal dedicated to publishing high-quality research in all areas of human genetics.
Related Collections
With Collections, you can get published faster and increase your visibility.
Genetic epidemiology in the era of biobanks and multi-omics: study designs, methods, and translation
Publishing Model: Hybrid
Deadline: Jul 31, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in