Wrangling a de novo sequencing benchmark
Published in Chemistry and Computational Sciences
The behind-the-paper story here is about just how iterative this process can be. In a sense, the first iteration of the benchmark is the version described in the initial DeepNovo paper, though for all I know that version itself represents multiple prior iterations. We used that benchmark in the first paper describing our Casanovo de novo sequencing model. When we began working on the second paper about Casanovo, we were motivated to create a new version of the benchmark because we weren’t sure how the database searching and FDR control were done.
The process, as outlined in my lab notebook, ended up producing ten versions of the benchmark.
- I started by manually downloaded the nine reference proteome fasta files from Uniprot. To produce the first version of the benchmark, I wrote scripts to automatically download all the mass spectrometry data using ppx, convert to MGF format using ThermoRawFileParser, and search the data using the Tide search engine followed by Percolator. The peptide sequences for peptide-spectrum matches (PSMs) accepted at a 1% PSM-level false discovery rate threshold were then inserted into the MGF files, discarding any spectra that failed to be
identified. - Initially, I used the same modifications that had been in the DeepNovo analysis. However, we realized that we should make this benchmark consistent with the set of modifications in MassIVE-KnowledgeBase, because those were the modifications we were training Casanovo with.
- For each of the nine species, I had initially used estimates of precursor m/z tolerance and fragment bin size generated by our tool, Param-Medic. However, it seemed more defensible to use the parameters listed in the publications describing these datasets, so I switched to using those.
- I discovered that in the initial download of the raw files, some of the downloads failed. So I fixed that problem and re-generated the benchmark.
- I had initially created a version of the benchmark with one big MGF file per species, but this was problematic because scan numbers ended up being repeated. So I switched to creating one MGF file per raw file.
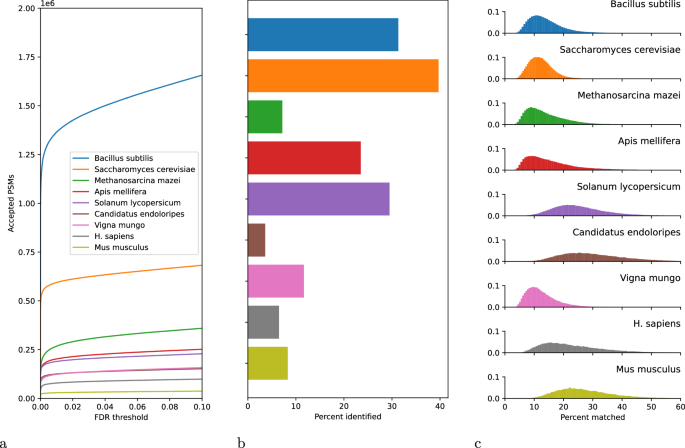
- We found that some of the annotations in the original DeepNovo benchmark did not properly account for isotope errors (see Figure 5 in this paper), so I added handling of isotope errors to my search parameters. Overall, this change did not make a big difference in the number of accepted PSMs. For one species (Vigna mungo) the number dropped slightly; for all the others it increased by a few thousand PSMs.
- We realized that some peptides in the benchmark were shared between species, so I added a post-processing step to eliminate these shared peptides.
- A user pointed out that our benchmark did not actually contain any N-terminal modifications. It turns out that this was a known bug in the Tide search engine, which had been recently fixed. I therefore re-ran the entire search procedure to generate a new version of the benchmark.
- One of the reviewers of the second Casanovo paper asked us to be sure that different modified forms of the same peptide sequence all be associated with a single species. This seemed like a good idea, so I made this change.
- Unfortunately, Tide and Casanovo do not agree on how to represent a peptide containing modifications: Tide puts them in square brackets, whereas Casnovo leaves off the brackets but precedes the mass with a “+”. I therefore added a cleaning step to convert all the Tide peptides to Casanovo format.
As this list makes clear, the process of creating the revised nine-species benchmark and ensuring its quality has been iterative. I fully expect to need to make additional updates to the benchmark as others begin using it, so I will continue to update our GitHub repository of scripts and the benchmark itself as needed.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Genomics in freshwater and marine science
Publishing Model: Open Access
Deadline: Jul 23, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in