A comprehensive AI model development framework for consistent Gleason grading
Published in Computational Sciences and General & Internal Medicine

AI-drive digital pathology is an emerging field that leverages artificial intelligence (AI) to enhance diagnostic processes. Despite its promise, the field grapples with critical challenges such as ensuring consistent performance across different hospitals, sample preparations and scanning equipment, and developing a dynamic platform that facilitates seamless interaction between pathologists and AI.
Our paper showcases Gleason grading in prostate cancer (PCa) to demonstrate a comprehensive workflow that is adaptable to all diagnostic model development relevant to digital pathology. The workflow comprises A!MagQC (image quality control), A!HistoClouds (cloud-based annotation), Pathologist-AI Interaction (PAI) for continuous model improvement. A standout feature of our study is the introduction of "image appearance migration" (IAM), a pioneering method designed to tackle the generalization and adoption challenges inherent in digital pathology.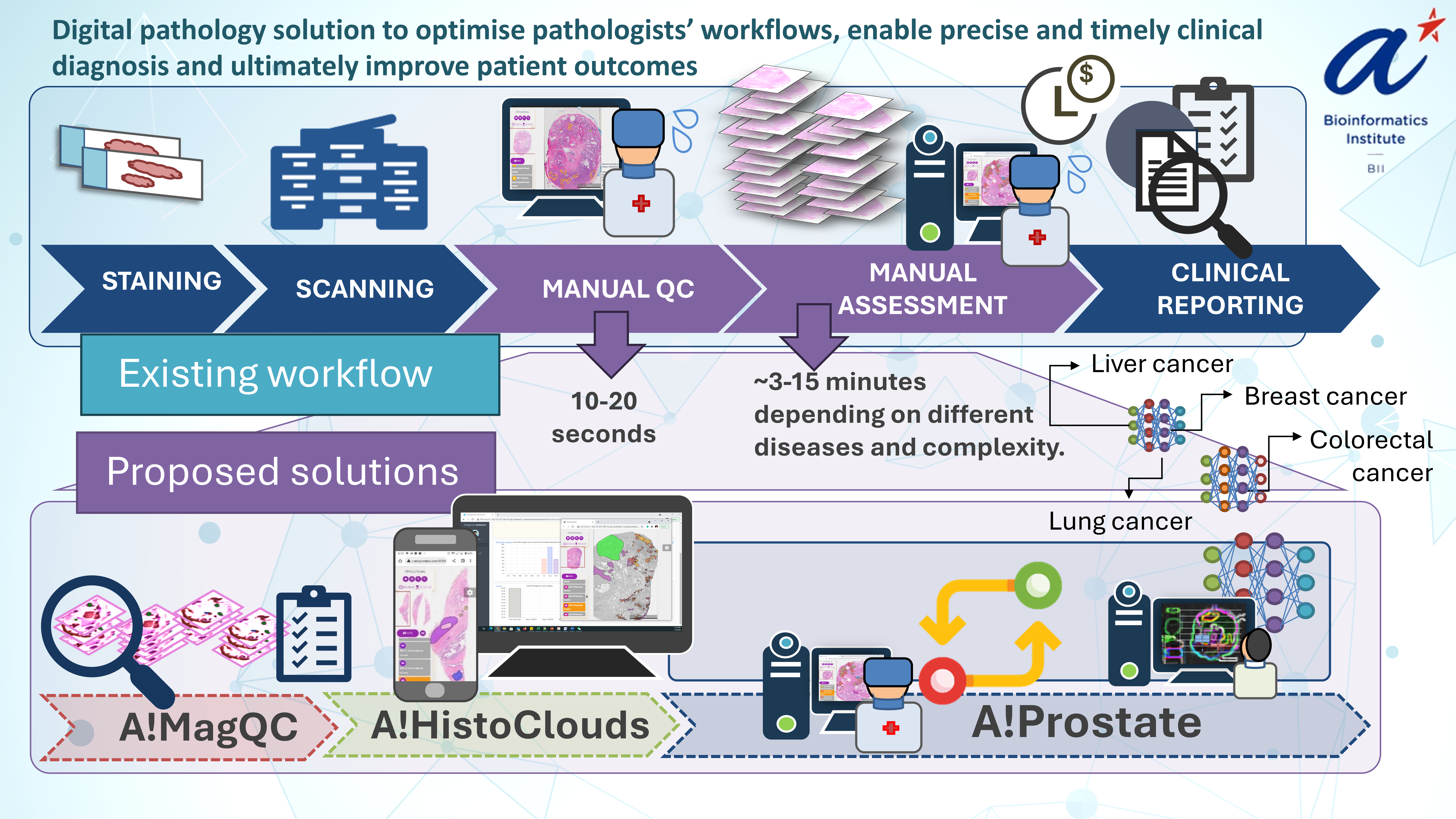
The Rise of Digital Pathology and AI's Impact
Digital pathology transforms traditional microscopic examination by digitizing pathological slides using scanners for enhanced accessibility and digital analysis. This technology facilitates the management and examination of slide images from anywhere, providing a substantial foundation for advanced diagnostic methods.
AI further enhances these capabilities by automating the analysis process, thus reducing the time pathologists need to spend on each case. It improves diagnostic precision, helps pathologists identify subtle patterns, and quantifies features more consistently and rapidly than traditional methods. This combination of digital pathology and AI not only improves accuracy but also significantly accelerates the workflow, enabling faster patient outcomes.
Challenges and Our Proposed Innovations in AI-Driven Pathology
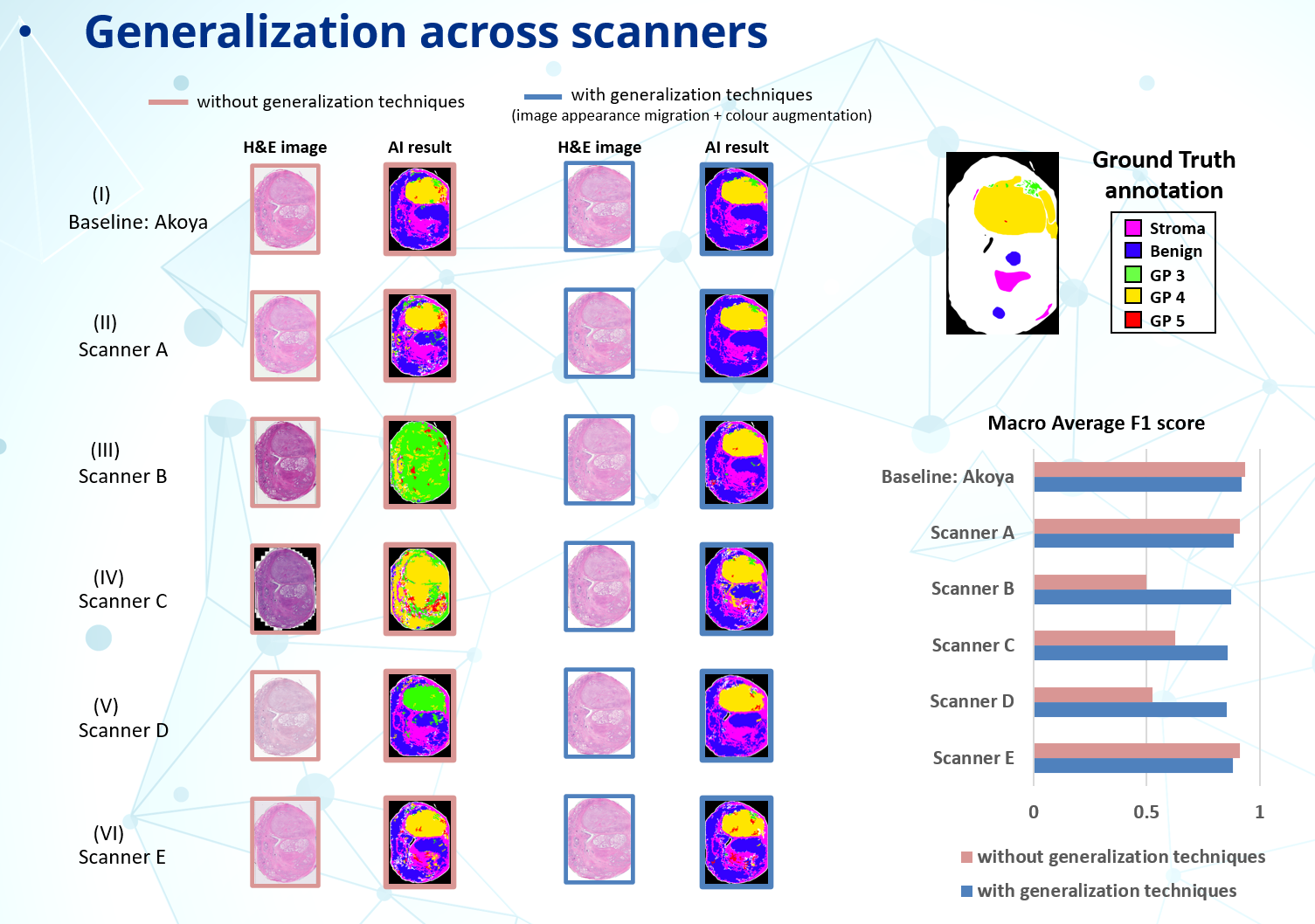
1. Generalization Challenges and Image Appearance Migration (IAM): Integrating AI into digital pathology requires addressing the challenge of ensuring robust generalization across varied datasets. Differences in scanning equipment and sample preparations can lead to significant image variations. These non-biological variations can drastically affect AI performance if unaddressed.
Our solution, IAM, effectively standardizes the appearance of images from various sources into a "reference space", ensuring consistent AI performance. This technique focuses on eliminating variations caused by non-biological factors (e.g., scanning, staining, etc.) while preserving critical biological and pathological differences for accurate AI analysis.
2. Creating High-Quality Image Datasets and Pathologist-AI Interaction: Quality control (QC) is important in traditional pathology using microscopes while it is still essential in digital era. Our approach automates this process with A!MagQC software, which identifies common quality issues in Whole Slide Images (WSI) and provides quantification results, thereby reducing the manual effort required for QC.
Bridging the gap between AI and pathologists, our model supports incremental learning, which enhances AI capabilities as new data is integrated. Semi-automatic annotations decrease the time and effort required by pathologists as model performance improves. This dynamic interaction is facilitated by the A!HistoClouds platform, closing the loop between AI improvements and pathologist input.
Data Sharing and Future Directions
We are committed to fostering a collaborative research environment. The AGGC2022 challenge (https://aggc22.grand-challenge.org/), part of MICCAI 2022, attracted over 250 teams, demonstrating widespread interest in addressing these challenges. Post-challenge, the dataset was made publicly available to encourage ongoing research. Our database was also cited in a recent paper from Harvard Medical School, published in Nature Medicine in March 2024 [1].
Looking forward, we plan to extend our innovative workflow to other cancer types, to further demonstrate the effectiveness of our proposed methods.
- Chen, R.J., Ding, T., Lu, M.Y. et al. Towards a general-purpose foundation model for computational pathology. Nat Med 30, 850–862 (2024). https://doi.org/10.1038/s41591-024-02857-3
Follow the Topic
-
Communications Medicine

A selective open access journal from Nature Portfolio publishing high-quality research, reviews and commentary across all clinical, translational, and public health research fields.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Healthy Aging
Publishing Model: Open Access
Deadline: Jun 01, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in