A cyan spie for salt in cells
Published in Chemistry

“To measure is to know.” is a quote from Lord Kelvin and it implies that measurements are the foundation of knowledge. But how can one measure anything in cells, which are tiny and transparent? First, you need a microscope. Second, you need some kind of paint to make the cells visible.
Coloring cells
The jellyfish Aequorea Victoria creates its own, natural pigment: the Green Fluorescent Protein (GFP). GFP emits green light, and has radically changed the way scientists look at cells. The DNA code that generates GFP is known and inserting the DNA with the code for GFP into cells is enough to make them shine green.

mTurquoise2, SYFP2 and mScarlet-I
This method can be used to stain all kinds of cells and even complete organisms, making them clearly visible with a microscope. Our lab at the University of Amsterdam has been developing and improving stains of different colors that are inspired by GFP. In this way, bright variants of cyan [1] and red fluorescent proteins [2] are created. These glowing colors all have their own, unique DNA code. The DNA can be shared with other labs and enables scientists to use these stains for their own research.
A sensitive GFP
The glowing proteins are mostly insensitive to their environment. But, by clever modification of the GFP, it is possible to make GFP-based stains that are sensitive to salt. These responsive stains are called biosensors. The classic biosensors emit little light at low salt concentrations and a lot of light at high salt concentrations. By measuring the amount of emitted light in cells with a microscope, changes in the concentration of salt can be followed in real-time. However, since the amount of emitted light also depends on the biosensor concentration, it is very difficult to accurately quantify salt concentrations based on the amount of emitted light.
A different view on fluorescence: lifetime
The green fluorescence that is emitted by GFP is preceded by the absorption of blue light. The time between the absorption and emission is the fluorescence lifetime. The fluorescence lifetime does not depend on the fluorescence intensity and is therefore an ideal parameter for measurements in cells. The fluorescence lifetime can be measured with a dedicated microscope, a Fluorescence Lifetime Imaging Microscope or FLIM. FLIM is very suited for accurate measurements, but there are hardly any biosensors with a lifetime that reports on the salt concentration.
Turquoise biosensors
The aim of the project of PhD candidate Franka van der Linden was to make a biosensor that uses fluorescence lifetime to measure the concentration of a specific salt, calcium. The new biosensor has a similar design as existing biosensors that use a 'circular permutated' variant of GFP [3]. But, instead of GFP, a cyan fluorescent protein named mTurquoise2 [1] was used.
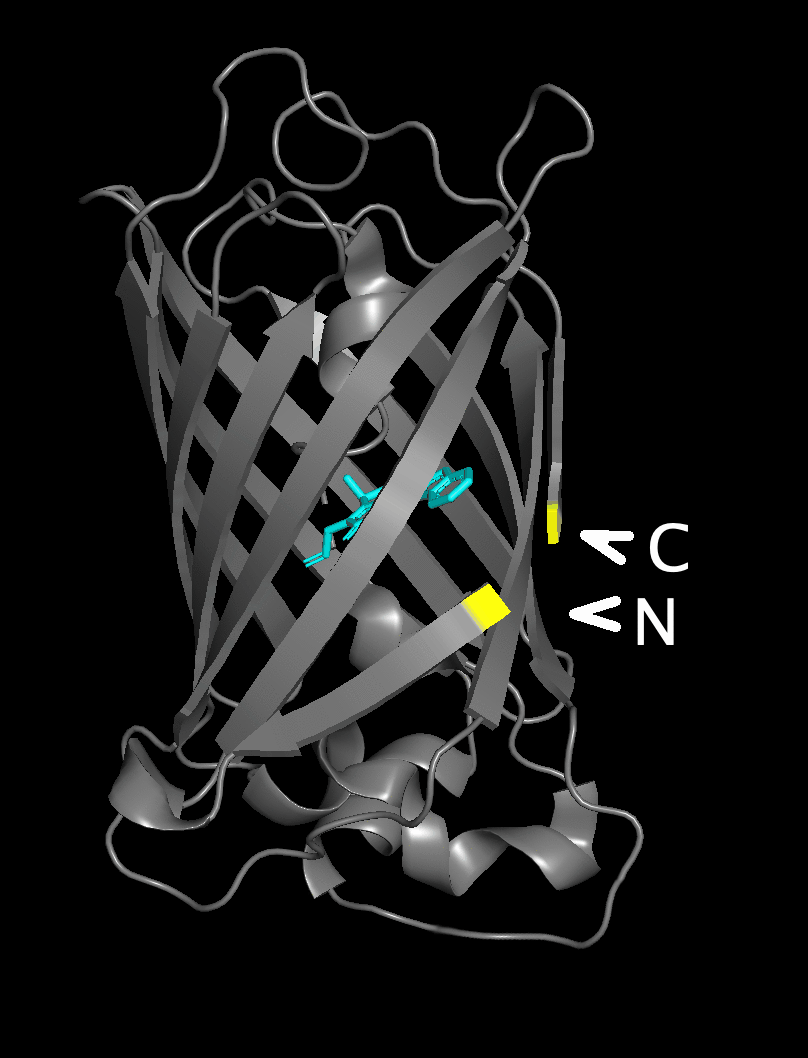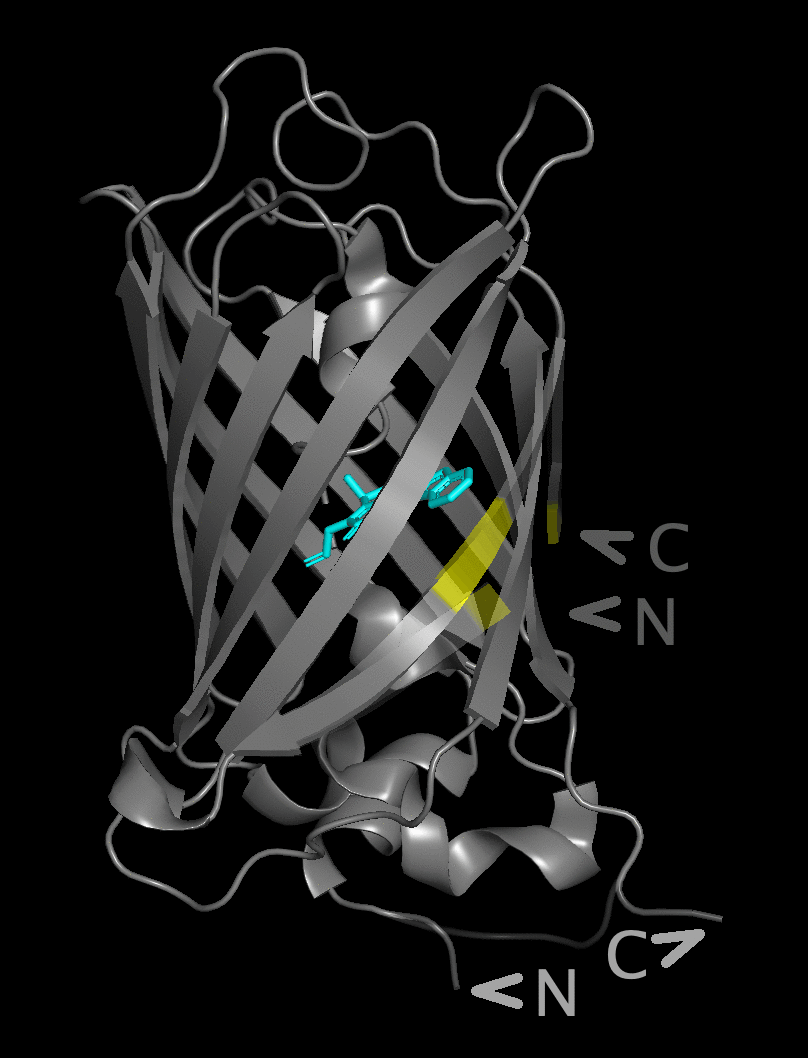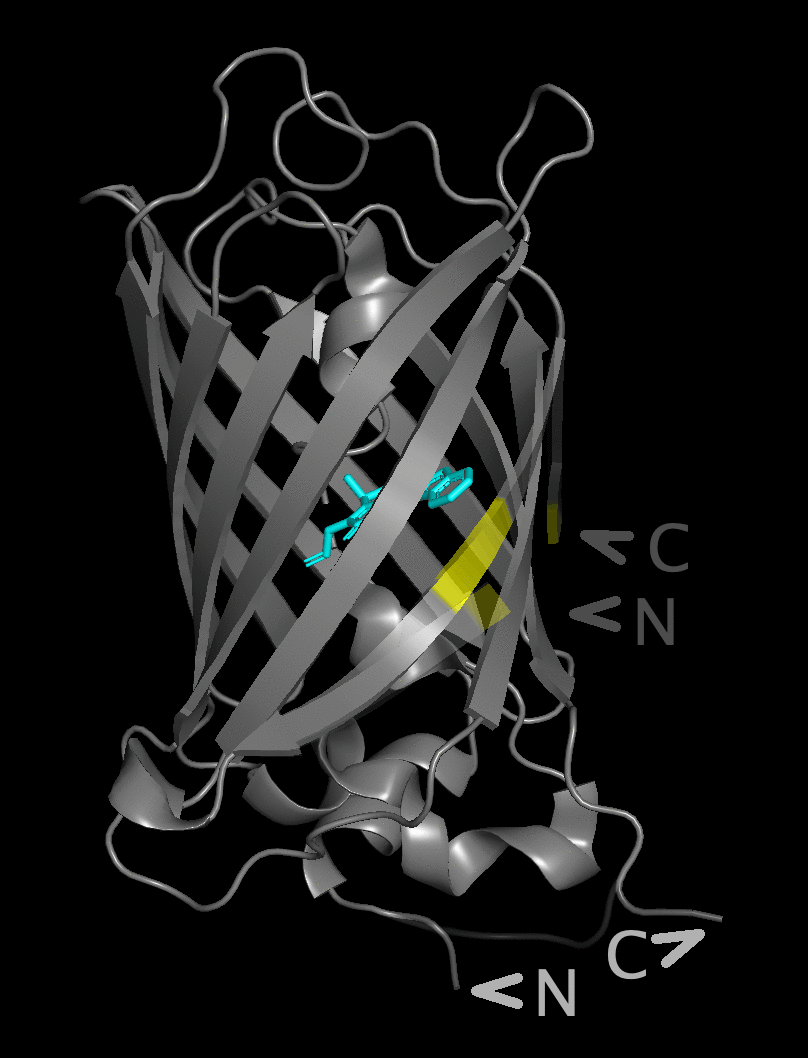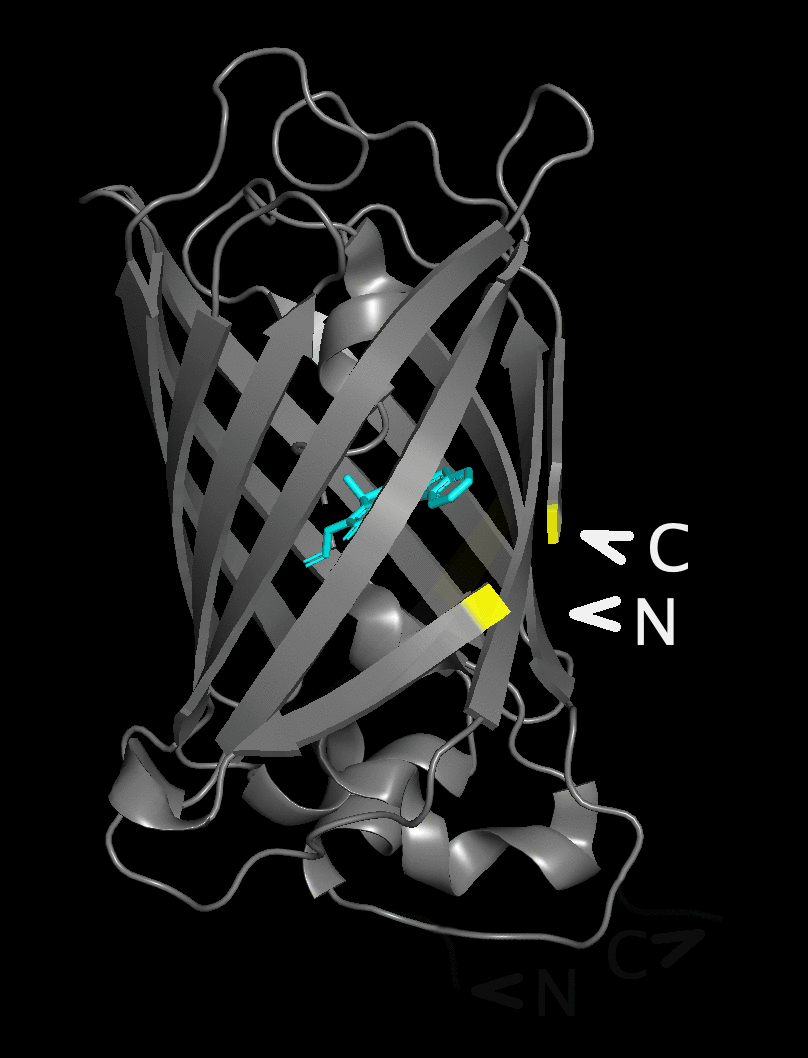
The figure flips between a structure with the native
N- and C-termini underneath the beta barrel and
a structure with the new position after circular permutation
in the yellow area near the chromophore.
After many months of making and trying different, slightly modified DNA codes, a biosensor was created that shows a lifetime change when calcium concentrations change. As with any measuring device, a careful calibration of the biosensor was needed before it could be properly used. After calibration, the fluorescence lifetime of the biosensor measured in cells can be converted into a calcium concentration.
A beneficial side-effect of using a cyan fluorescent protein is that the biosensor is not sensitive to acid. Whereas GFP-based biosensors lose their fluorescence when the pH drops, the cyan biosensor is not affected by pH changes. The lack of pH sensitivity is another reason why this sensor is suitable for quantitative measurements of calcium concentrations. The results are published recently in Nature Communications [4] and plasmids with DNA that encode for the biosensor are available from addgene.
To measure calcium, is to know calcium.
Calcium ions play a key role in different cellular processes. For example, calcium ions are needed for the contraction of muscles, calcium is involved in brain activity and calcium ions are released after the perception of hormones by cells. Given the universal role of calcium in cellular processes, scientists are interested in measuring it. To demonstrate the application of the new biosensor, it was used to detect the increase of the calcium concentration in endothelial cells by stimulation with histamine.
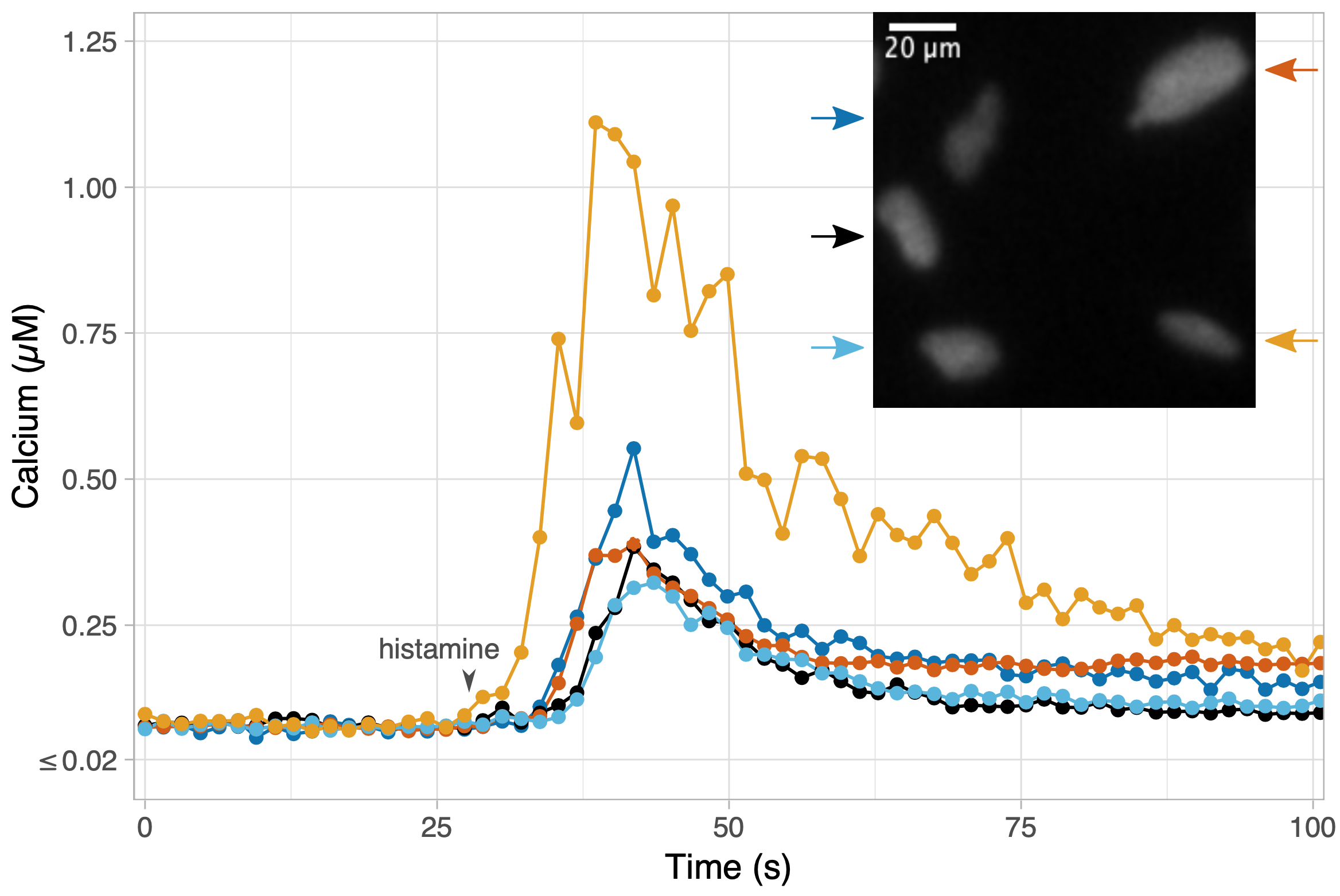
In addition, the calcium sensor was used in collaboration with Eike Mahlandt, Janine Arts and Jaap van Buul (Sanquin Research, Amsterdam), to detect calcium concentration in a layer of endothelial cells, mimicking the inner layer of blood vessels. It was observed that calcium levels hardly change when white blood cells pass the layer of endothelial cells.
Another demonstration of the biosensor was done in collaboration with Joep Beumer, Jens Puschhof and Hans Clevers (Hubrecht Institute, Utrecht). The biosensor was used to detect calcium in organoids. The organoid is a mini-gut that responds like a natural gut. The calcium dynamics were precisely quantified showing the potential of the new biosensor as a new and accurate measurement device.
References
[1] Goedhart, J., D. von Stetten, M. Noirclerc-Savoye, M. Lelimousin, L. Joosen, M.A. Hink, L. van Weeren, T.W.J. Gadella, and A. Royant. 2012. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nature Communications. 3:751. doi:10.1038/ncomms1738.
[2] Bindels, D.S., L. Haarbosch, L. van Weeren, M. Postma, K.E. Wiese, M. Mastop, S. Aumonier, G. Gotthard, A. Royant, M.A. Hink, and T.W.J. Gadella. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nature Methods. 14:53–56. doi:10.1038/nmeth.4074.
[3] Nasu, Y., Y. Shen, L. Kramer, and R.E. Campbell. 2021. Structure- and mechanism-guided design of single fluorescent protein-based biosensors. Nature Chemical Biology. 17:509–518. doi:10.1038/s41589-020-00718-x.
[4] van der Linden, F.H., E.K. Mahlandt, J.J.G. Arts, J. Beumer, J. Puschhof, S.M.A. de Man, A.O. Chertkova, B. Ponsioen, H. Clevers, J.D. van Buul, M. Postma, T.W.J. Gadella, and J. Goedhart. 2021. A turquoise fluorescence lifetime-based biosensor for quantitative imaging of intracellular calcium.. Nature Communications. 12:7159. doi: 10.1038/s41467-021-27249-w
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in