A deep-learning approach to predict spatial cellular architecture and prognosis in glioblastoma from histology images
Published in Cancer

Glioblastoma (GBM) is the most aggressive tumor type in the human central nervous system. Despite recent advancements in neurosurgery, the effectiveness of chemotherapy and radiotherapy is still limited1. The therapeutic resistance results from intra-tumoral heterogeneity and cell-state plasticity2,3. In the past decade, our understanding of cellular heterogeneity in GBM has deepened resulting from the advancements of high-throughput technologies such as single-cell RNA sequencing and spatial transcriptomics. However, despite their potency, current spatial transcriptomics and proteomics assays suffer from two major limitations. First, they are still expensive, require specialized expertise, and are not yet integrated into routine assays for cancer diagnosis, which limits their clinical applications. Second, the input tissue size for analysis is much smaller than the actual tumors, potentially introducing bias towards the tested tumor sections. For instance, the 10X Visium platform allows profiling of tissue sections up 6.5 x 6.5 mm or 11 x 11 mm in size4. Nevertheless, the diameter of clinical GBM cases can range from 0.1 cm to 10 cm5. These limitations have posed challenges to our understanding of the intricate associations between cell-type compositions, spatial cellular organization, and patient prognosis.
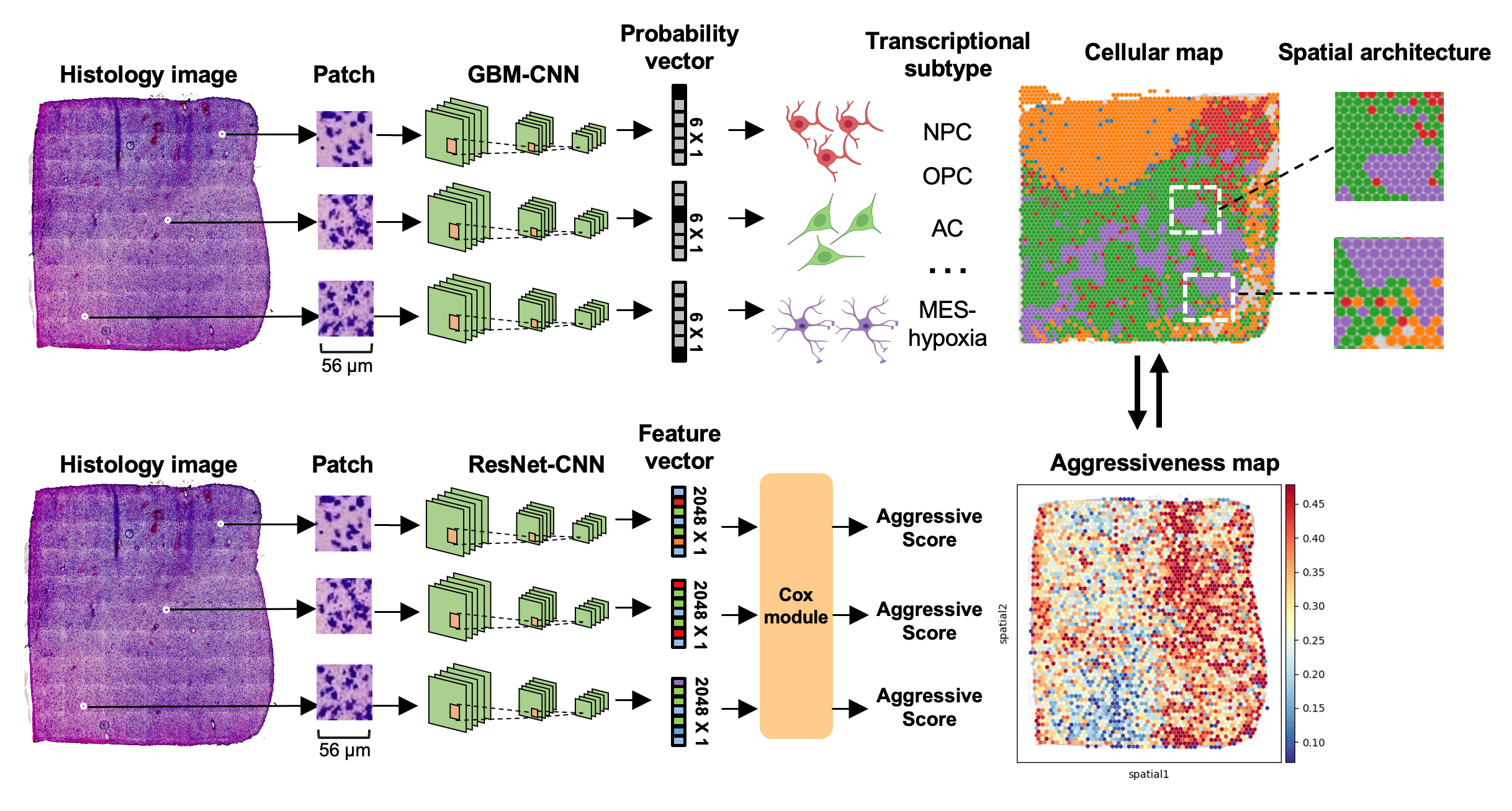
A deep-learning framework that predicts spatial cellular architecture and prognosis in glioblastoma.
Through a collaborative effort by scientists from Stanford University, a machine-learning framework is developed, which leverages H&E-stained histology images to infer the spatial cellular architecture and prognosis in GBM. This framework comprises two deep-learning models. The first model, called GBM-CNN, utilizes histology images to infer the transcriptional subtypes of GBM cells. The study considers five known transcriptional subtypes, namely (1) neural-progenitor-like (NPC-like), (2) oligodendrocyte-progenitor-like (OPC-like), (3) astrocyte-like (AC-like), and (4) mesenchymal-like (MES-like). The MES-like subtype is further classified into hypoxia-dependent (MES-hypoxia) and hypoxia-independent (MES-like) groups based on the expression of hypoxia-response genes (e.g., HILPDA, VEGFA) and glycolytic genes (e.g., GAPDH, LDHA)6.
To train GBN-CNN, the authors integrate four spatial transcriptomics datasets7–9, encompassing a total of 75,625 tissue spots comprising both GBM and normal brain tissues. The model is evaluated using internal and external validations. Results show that for predicting tumor cells, the F1 score is 0.86 with a standard deviation (SD) of 0.15. The area under the receiver operating characteristic curve (AUROC) reaches 0.93 (SD: 0.05). For predicting T cells, the sensitivity is 0.80 (SD: 0.08), specificity is 0.83 (SD: 0.05), and AUROC is 0.80 (SD: 0.08). For predicting macrophages, the sensitivity is 0.84 (SD: 0.09), specificity is 0.81 (SD: 0.07), and AUROC is 0.89 (SD: 0.11). These results demonstrate that the GBM-CNN is able to accurately predict the subtypes of tumor cells and the presence of immune cells from histology images.
Leveraging this model, the authors phenotypically analyze 40 million tissue spots from 410 patients across two independent cohorts. The analysis led to the identification of consistent associations between tumor architecture and prognosis. Notably, patients with poor prognosis exhibit higher proportions of tumor cells expressing a hypoxia-induced transcriptional program. Furthermore, a clustering pattern of astrocyte-like tumor cells is associated with worse prognosis, while dispersion and connection of the astrocytes with other transcriptional subtypes correlate with a decreased risk.
To further identify survival-associated transcriptional programs, the authors develop a separate deep-learning model that utilizes histology images to predict prognosis. The model aims at assigning an aggressive score to each tumor region, with high aggressive scores contributing to a worse prognosis. Applying the model to spatial transcriptomics data reveals survival-associated regional gene expression programs. Genes related to injury response and glycoprotein metabolism are significantly upregulated in tumor cells with higher aggressiveness.
Finally, the study develops a user-friendly software, named GBM360 (gbm360.stanford.edu), that allows users to characterize tissue compositions and spatial cellular organization of new GBM cases. GBM360 enables three functionalities (1) predict transcriptional subtypes of tumor cells from histology images and generate high-resolution spatial cellular maps, (2) quantitively characterize the spatial distribution of malignant cells using three types of spatial statistics, and (3) predict prognosis and visualize regional aggressive scores.
The approach presented by the authors utilizes histology images, a widely available and easy-to-obtain diagnostics assay, to unravel the spatial cellular heterogeneity within GBM. The results have the potential to improve our understanding of how tumor cells are spatially organized and interact with their immediate microenvironment. Using the presented approach, the authors discovered the associations between spatial cellular organization and patient prognosis, which can lead to the improvement in personalized diagnosis and treatment.
- Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
- Larsson, I. et al. Modeling glioblastoma heterogeneity as a dynamic network of cell states. Mol. Syst. Biol. 17, e10105 (2021).
- Yabo, Y. A., Niclou, S. P. & Golebiewska, A. Cancer cell heterogeneity and plasticity: A paradigm shift in glioblastoma. Neuro. Oncol. 24, 669–682 (2022).
- Williams, C. G., Lee, H. J., Asatsuma, T., Vento-Tormo, R. & Haque, A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 14, 68 (2022).
- Kozak, K. R. & Moody, J. S. Giant cell glioblastoma: a glioblastoma subtype with distinct epidemiology and superior prognosis. Neuro. Oncol. 11, 833–841 (2009).
- Neftel, C. et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835-849.e21 (2019).
- Ravi, V. M. et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 40, 639-655.e13 (2022).
- Ren, Y. et al. Spatial transcriptomics reveals niche-specific enrichment and vulnerabilities of radial glial stem-like cells in malignant gliomas. Nat. Commun. 14, 1028 (2023).
- Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in