A machine-learning approach to phylogenetic tree inference
Published in Ecology & Evolution
Reconstructing a phylogenetic tree for a group of organisms based on molecular sequence data is a fundamental challenge in evolutionary research. When only a few dozen of species are analyzed, billions of alternative phylogenetic trees could potentially describe the evolutionary patterns, thus rendering the search for the tree that best describes the data algorithmically challenging.
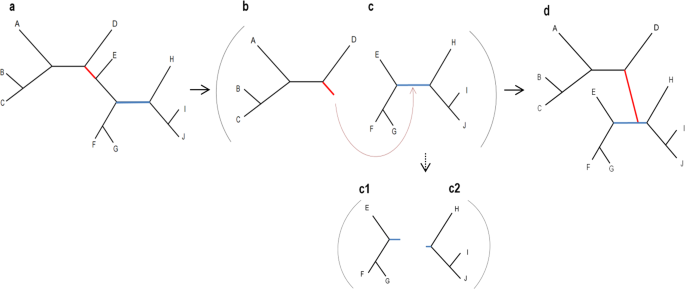
If one were to develop a naïve-search algorithm, they would possibly start with a candidate tree and generate all its immediate neighboring trees by pruning each branch in turn and regrafting it to some other branch in that tree. Then, the same procedure would be done with the highest-scoring neighboring tree as the current candidate tree. These steps should be computed iteratively until convergence. As the number of possible tree topologies increases super-exponentially with the number of sequences, previously developed heuristic strategies attempt to balance between accuracy and running time. That means that providing a feasible solution comes at the cost of accuracy.

The challenge: speed up heuristic searches without compromising accuracy.
Our solution: harness machine learning to boost heuristic tree searches.
How: we trained a machine-learning algorithm to rank the candidate trees according to their propensity to improve the fit to the data, without actually calculating it.
To this end, we generated a starting tree and all its immediate neighboring trees for each of the 4,200 empirical datasets we collected (resulting in dozens of millions training samples). For each possible move to a neighboring tree, we extracted 19 features that represent that move, and for each potential neighboring tree, we computed the increase/decrease in the fit to the data. At this point, we were ready to train a machine-learning algorithm that would predict the change in the fit, according to these features. Our trained-random-forest-regression model is able to rapidly predict which are the most promising candidate trees, and which can be discarded. This way we avoid the computationally intensive evaluation of many trees.
Take-home message
There are patterns in the data that can be learned using a machine-learning model. More generally, we provided a proof of concept that learning approaches can greatly improve our ability to accurately and efficiently reconstruct phylogenetic trees.
---------
Stay tuned! We are already progressing with this research direction to provide improved AI-based algorithms for phylogeny reconstruction.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in