A step towards improving anti-C. difficile bacterial therapeutics: Identifying interactions that are more robust across strains and environmental contexts
Published in Microbiology, Protocols & Methods, and Biomedical Research
Although antibiotics can be used to treat Clostridioides difficile infection (CDI), they kill the good bacteria in our gut and perturb the balance of the microbiome. This results in the loss of colonization resistance and increases the chance of recurrence of CDI (rCDI). Fecal Microbiota Transplantation (FMT) has emerged as another treatment option, where fecal samples from healthy donors are transplanted into sick patients. Although FMT has high success rates for treating rCDI, it is generally regarded as unreliable since we do not know the exact identity of the microbes (i.e., black box). This leads to safety concerns such as the transfer of antibiotic-resistant bacteria or other pathogens, and other possible side effects.
The recent years have seen substantial progress in developing defined bacterial therapeutics that are optimized to inhibit C. difficile, such as the 8-member defined consortia from Vedanta Biosciences (VE303). This live bacterial therapeutic (LBT) has passed phase 2 clinical trial and is currently undergoing phase 3 1. However, defined communities have yet to realize the remarkable efficacy seen for FMT bacterial therapeutics 2,3, suggesting that there are critical properties that enable certain human gut microbiomes to inhibit C. difficile that we do not yet understand. Since defined communities have substantially lower species richness and diversity than FMT samples, we hypothesize they are less robust to environmental variability due to reduced functional redundancy 4. However, increasing the number of species in a live bacterial therapeutic is undesirable since the community size is currently proportional to the manufacturing time and costs. We hypothesize that the lack of robustness of defined bacterial therapeutics to environmental variability may limit their therapeutic potential. Sources of variability that a LBT could encounter in the human gut microbiome include strain-level genetic and phenotypic diversity of human gut pathogens, inter and intra-individual variation in gut microbiome compositions, host genetics, immune responses, and diet.
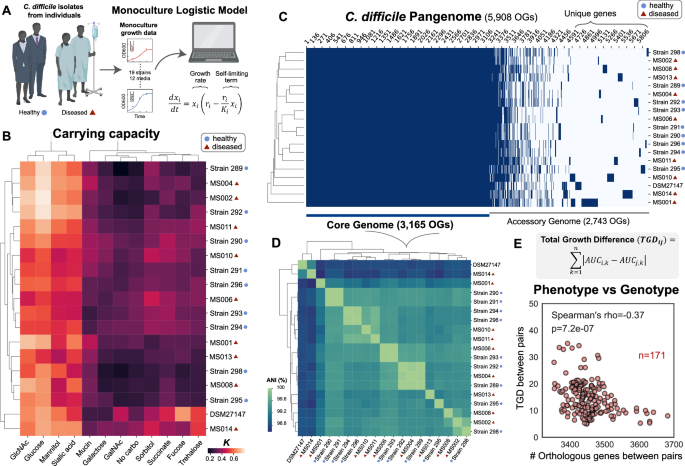
In this study, we tackled how inter-species interactions of C. difficile with human gut microbiota varied across two major environmental factors, C. difficile strains and nutrient environments. Species and communities that can inhibit multiple C. difficile strains across different nutrient environments may display enhanced robustness to environmental variability. The increase in abundance of certain resources in a perturbed gut following antibiotic treatment has been shown to promote the growth and colonization of C. difficile 5, indicating that nutrient variations could affect inter-species interactions among C. difficile and gut bacteria. Further, C. difficile has extreme genetic variability, comprising hundreds of strain types that are distributed across at least 8 phylogenetic clades. This species is defined by a large pangenome with high variation in the accessory genomes, leading to variation in the regulation of metabolic pathways and virulence. Notably, rCDI is not always due to infection with the same strain 6, and thus the degree of colonization resistance for an individual may depend on the specific C. difficile strain. Although the ecology of the human gut microbiome is a critical determinant of C. difficile colonization, the contribution of C. difficile strain-level variability to these interactions is currently unknown.
Our collaborator, Professor Nasia Safdar, works at the University of Wisconsin-Madison Hospital and her research explores the effects of interventions (e.g. diet, probiotics, and FMT) to prevent and reduce healthcare-associated infection including CDI, with some ongoing clinical trials in this area. Using C. difficile isolates from different subjects (healthy and diseased individuals) from her lab, we used a bottom-up approach that combines high-throughput community assembly with computational modeling to map inter-species interactions of genetically diverse C. difficile strains in different nutrient environments (Figure 1). Guided by our model, we identified certain anti-C. difficile communities that are robust to variation in C. difficile genetics and resource environments. Identifying optimized anti-C. difficile communities that could be effective across environmental contexts would be challenging using traditional screening and top-down approaches previously used to identify defined communities with desired functions. In addition to functional redundancy, other mechanisms that could promote the robustness of anti-C. difficile activity to environmental variability includes identifying species that maximize metabolic niche overlap with C. difficile or communities that possess multiple mechanisms that inhibit C. difficile including resource competition and antimicrobial activities.
.png)
Figure 1. By using a bottom-up approach that combines high-throughput experiments and computational modeling, we investigate how inter-species interactions between C. difficile and gut microbiota vary across strain variability and nutrient landscapes.
Using our workflow, we found that C. difficile growth was frequently inhibited by gut bacteria in media that promotes high resource competition, but most human gut species cannot inhibit C. difficile in media that mimics post-antibiotic perturbation where there are multiple preferred carbohydrates for C. difficile. This indicates that nutrient environments play a major role in shaping C. difficile growth in communities. Future studies could explore how human diets (e.g. dietary fibers) affect inter-species interactions of C. difficile and gut bacteria. In addition, we observed that C. difficile strains exhibit different degrees of competition for proline to perform Stickland metabolism with Clostridium scindens, revealing how metabolic variation across C. difficile strains could lead to differential metabolic niches and alterations in inter-species interactions in human gut communities. Our results highlight the importance of considering robustness to strain variations and nutrient environments as a design factor for anti-C. difficile consortia.
In addition to C. difficile growth, toxin production is a major determinant of C. difficile’s ability to cause infection, but we lack an understanding of how toxin production is shaped by diverse human gut species. By characterizing C. difficile toxin expression in the presence of human gut communities, we showed a lack of correlation between C. difficile toxin production and growth-mediated inter-species interactions, suggesting that inhibiting C. difficile growth may not always protect against CDI unless C. difficile is excluded from the community. Thus, the identification of C. difficile inhibitors should consider both inhibition of growth and toxin production.
Our approach identified Clostridium hiranonis as a “universal” C. difficile growth and toxin inhibitor that is robust to variation in C. difficile strains and nutrient environments both in vitro and in the mammalian gut. C. hiranonis has a high metabolic niche overlap with C. difficile and is also able to consume amino acids via the Stickland metabolism similar to C. difficile. We reveal that C. hiranonis blocks access of C. difficile to alternative resource niches and leads to a massive metabolic alteration in C. difficile, which in turn impacts toxin production. Consistent with in vitro results, C. hiranonis robustly ameliorated the disease severity of a C. difficile challenge in a murine model in response to two different C. difficile strains and variations in nutrient environments. Further, the inhibitory effects of C. hiranonis on C. difficile can be augmented with the addition of specific human gut bacteria. Future studies could test the protective ability of different defined communities containing C. hiranonis against C. difficile in the mammalian gut. Although there is no evidence regarding the role of C. hiranonis on CDI outcomes in humans, the presence of C. hiranonis is negatively associated with C. difficile colonization in dogs and cats 7.
Overall, we showcase our quantitative systems-biology approach to map context-dependent interactions and provide insights into the mechanisms that could enhance the robustness of inhibition across strains and environments. We demonstrate that interactions that lead to global shifts in metabolism and other cellular processes may exhibit greater robustness to environmental variability. More broadly, this framework that considers robustness as a feature could be applied to the design of anti-pathogen bacterial therapeutics beyond C. difficile.
References
1. Dsouza, M. et al. Colonization of the live biotherapeutic product VE303 and modulation of the microbiota and metabolites in healthy volunteers. Cell Host & Microbe 30, 583-598. e588 (2022).
2. Sims, M. D. et al. Safety and tolerability of SER-109 as an investigational microbiome therapeutic in adults with recurrent Clostridioides difficile infection: a Phase 3, open-label, single-arm trial. JAMA Network Open 6, e2255758-e2255758 (2023).
3. Louie, T. et al. VE303, a defined bacterial consortium, for prevention of recurrent Clostridioides difficile infection: a randomized clinical trial. JAMA 329, 1356-1366 (2023).
4. Lee, K. K., Park, Y. & Kuehn, S. Robustness of microbiome function. Current Opinion in Systems Biology, 100479 (2023).
5. Jenior, M. L., Leslie, J. L., Young, V. B. & Schloss, P. D. Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. MSystems 2, 10.1128/msystems. 00063-00017 (2017).
6. Johnson, S., Adelmann, A., Clabots, C. R., Peterson, L. R. & Gerding, D. N. Recurrences of Clostridium difficile diarrhea not caused by the original infecting organism. The Journal of infectious diseases 159, 340-343 (1989).
7. Takáčová, M., Bomba, A., Tóthová, C., Micháľová, A. & Turňa, H. Any future for faecal microbiota transplantation as a novel strategy for gut microbiota modulation in human and veterinary medicine? Life 12, 723 (2022).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in