Accelerating Drug Discovery with Machine Learning for Targeted Protein Degraders
Published in Chemistry and Computational Sciences

Introduction
Targeted protein degraders (TPDs) have emerged as a promising new modality in drug discovery. These molecules are able to selectively degrade disease-causing proteins and have the potential to tackle diseases that were previously considered "undruggable". However, the application of machine learning (ML) models for TPDs’ property predictions has been limited and questioned. In our recent paper titled "Application of machine learning models for property prediction to targeted protein degraders", we demonstrate the suitability of ML-based quantitative structure-property relationship (QSPR) models for predicting various properties of TPD molecules. Our work sheds light on the potential of ML in accelerating drug discovery for TPDs.
Comprehensive Evaluation of ML for TPDs
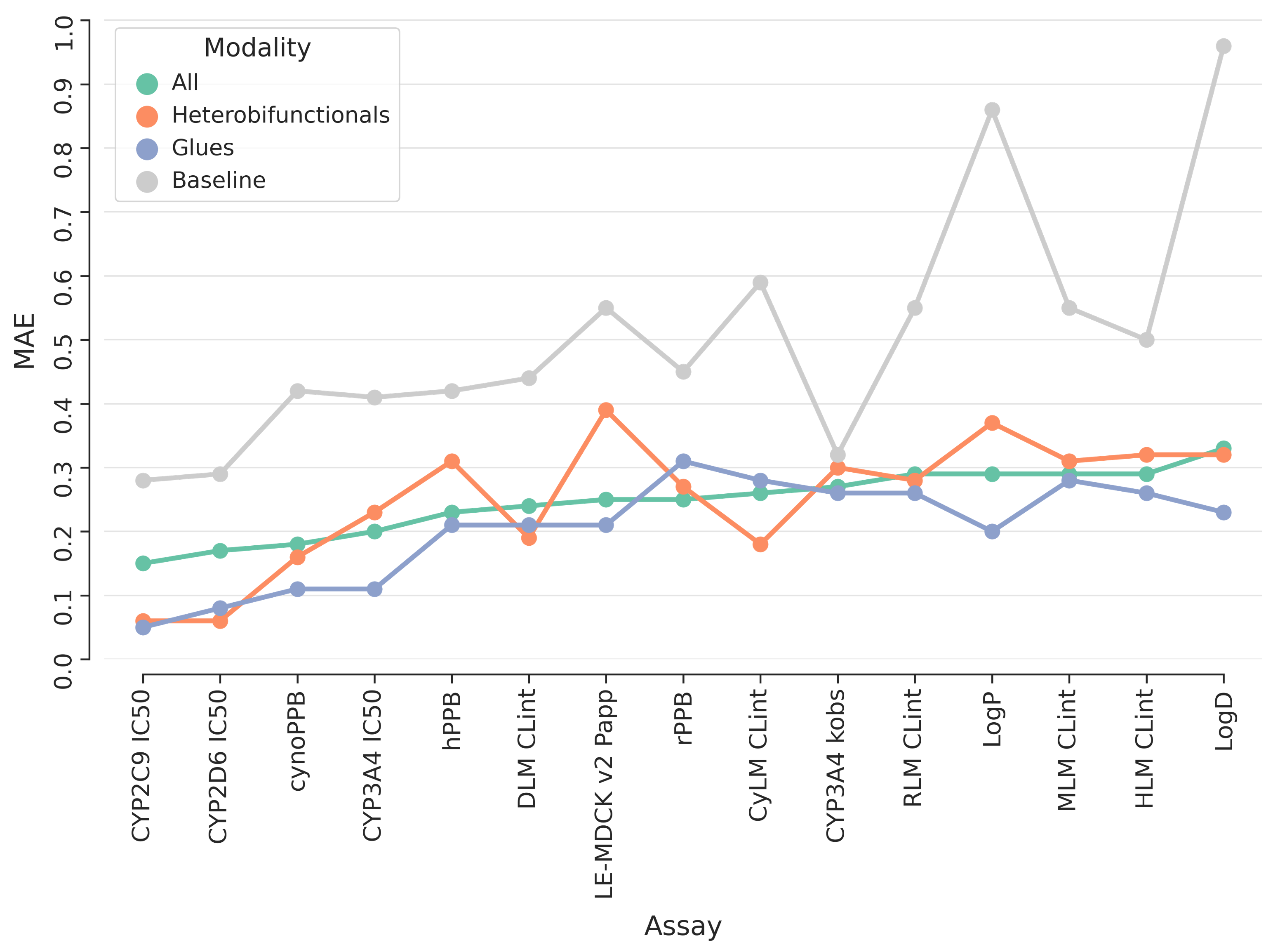
Our study evaluated ML models for the prediction of ADME (absorption, distribution, metabolism, and excretion) and physicochemical properties of TPDs, including both molecular glue and heterobifunctional submodalities. We developed and tested ML models using existing experimental data, accurately predicting relevant properties of TPDs, such as passive permeability, metabolic clearance, or lipophilicity. Surprisingly, the performance of these ML models on TPDs was comparable to that of other modalities, with the molecular glue submodality showing the lowest prediction errors.
Challenges and Refinements
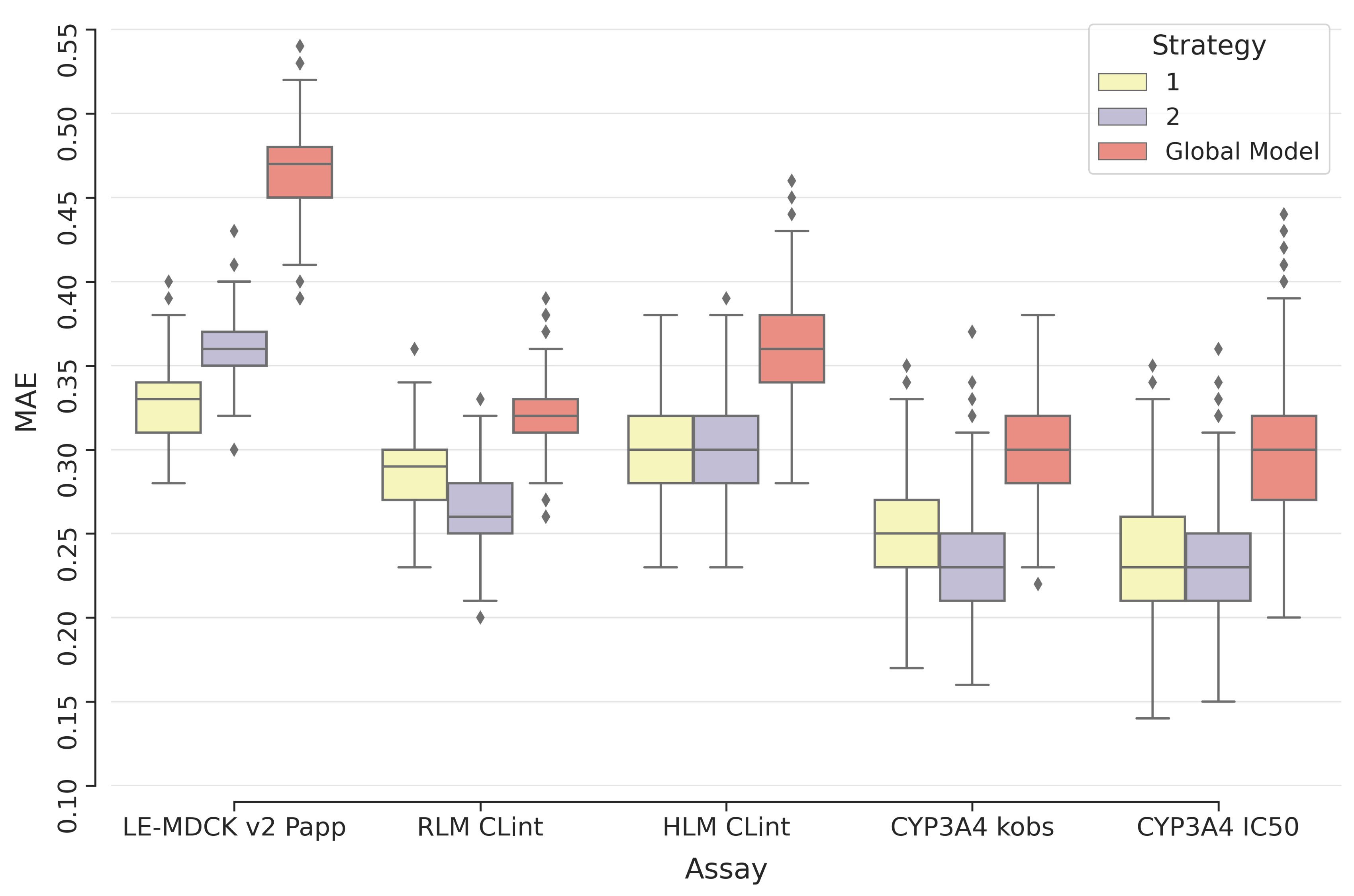
Our results revealed that predictions for heterobifunctional TPDs remain more challenging. However, the implementation of transfer learning strategies, such as fine-tuning models with heterobifunctional TPDs’ data, improved predictive performance across different ADME endpoints. This highlights the possibility for further refinement and improvement as more data becomes available.
The Potential of Surrogate Datasets
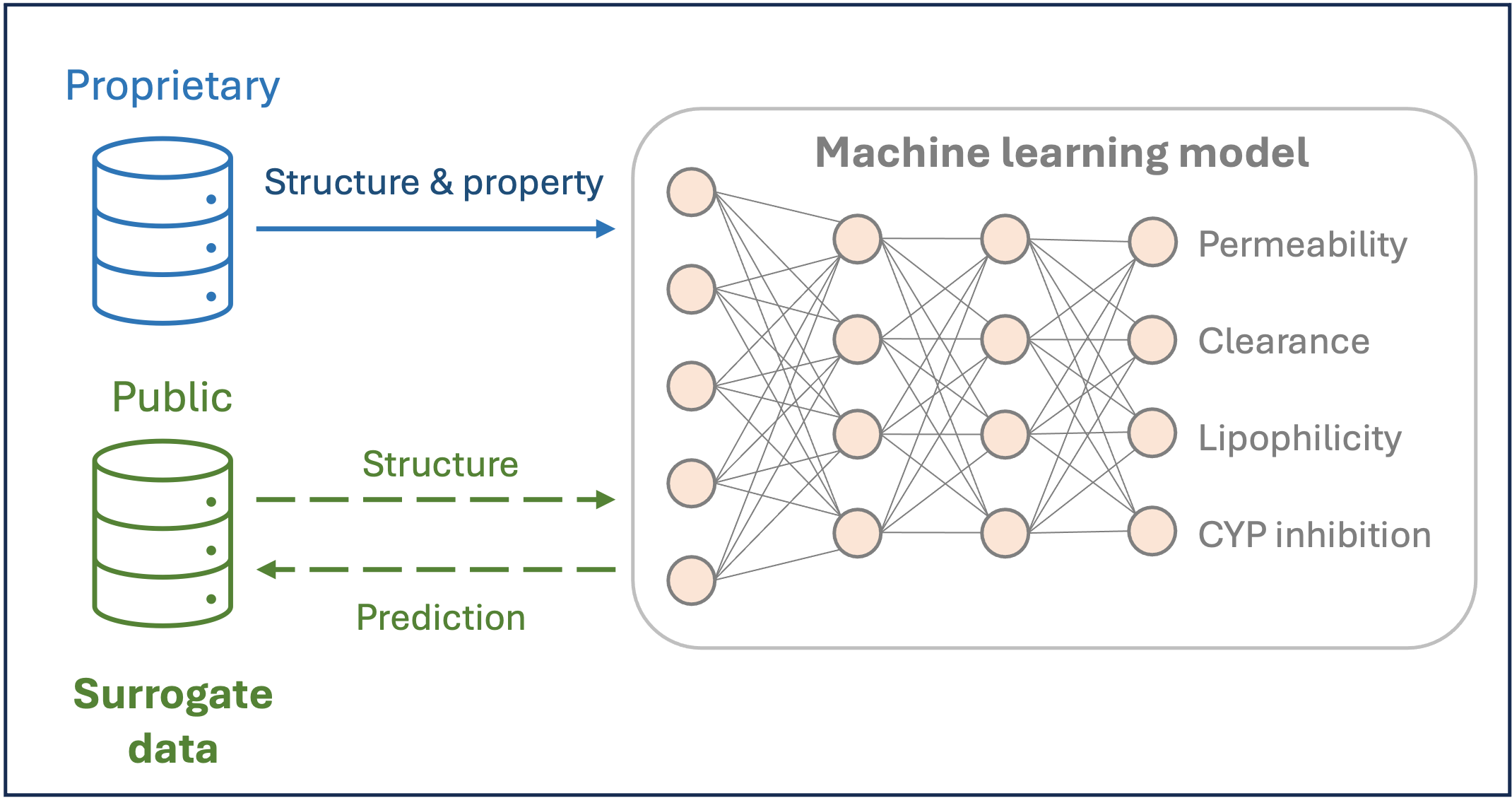
Our study provides a surrogate dataset with over 270,000 structures, annotated with in-house model predictions for twenty-five molecular properties. We showcased the potential of using ML-based QSPR models with surrogate data. Hence, this dataset offers exciting prospects for the advancement of ML models in the public domain.
Implications for Pharmaceutical Research
The integration of ML models into the design-make-test-analyze (DMTA) cycle of drug discovery has already proven beneficial in prioritizing compound ideas and experiments. However, the use of ML models for TPDs has been relatively limited compared to traditional modalities. Our findings shed light on the applicability of ML to TPDs and have implications for pharmaceutical research. Specifically, ML models show promise of accelerating the design of TPDs with favorable ADME properties and thus should be encouraged in TPD programs.
Conclusion
The ability of ML models to predict ADME properties for TPDs, combined with the potential for improvement through transfer learning, could enable more efficient drug design and advance the field of TPDs in drug discovery. As the availability of data for TPDs continues to increase, including with surrogate datasets, additional modeling strategies can be explored, opening up opportunities to more accurately predict TPDs’ properties based on molecular structures.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in