Monocots are one of the most numerous, ecologically significant, and economically significant lineages of land plants1, with more than 85,000 species and 21% of the world’s plants. Furthermore, monocots are renowned for their distinctive morphological traits, inhabiting a variety of habitats with extraordinarily rich terrestrial development patterns, and directly or indirectly providing the primary source of human sustenance in the form of grains or food crops such as rice, wheat, and maize. Therefore, botanists and evolutionary biologists have always been interested in understanding the cause and pattern of morphological evolution, geographic diversity, and ecological adaption of monocots.
Monocots can be categorized into 77 families and 12 orders according to morphological and genetic information2,3. In contrast to other angiosperms, monocots only have one cotyledon in the embryo, and their stem’s vascular bundles are dispersed in the shape of stars, with only primary phloem present and no cambium or secondary xylem. The Acorales are related to all other monocots as siblings, with only one family, Acoraceae1, and just one genus, Acorus. Comparative analysis with other angiosperms can show the molecular process of major trait innovation in the creation and evolution of monocots, including vascular cambium, secondary xylem, and cotyledon development, due to its unique evolutionary location. Acorus species also have a complicated evolutionary history4-7. Although officially only two species - with three varieties - have been accepted by Plants of the World Online5, four to five species and a few dozen varieties have been suggested in Acorus4. The two accepted species, i.e., A. gramineus Solander ex Aiton and A. calamus Linnaeus, are confined to the humid areas of temperate, tropical, and subtropical Asia and North America6.

Figure 1. A. calamus and A. gramineus in natural habital. a. A. calamus grow in wet land. b. Flowering plant of A. gramineus. c. Flowering plant of A. gramineus. d. A. gramineus grow on the rock.
A. gramineus is diploid 2n = 2x = 24 and A. calamusis tetraploid 2n = 4x = 44. It was discovered that A. gramineus had up to 60% of AB-type K-mer, indicating that it is a diploid with a genome size of 409.66 Mb. A. calamus is an allotetraploid, as evidenced by the larger percentage of AABB-type K-mer pairs (43% AABB type vs 23% AAAB type), and the size of the two subgenomes was 348.65 Mb. The complete Benchmarking Universal Single-Copy Orthologs (BUSCO)8 genes in the genome of diploid A. gramineus was only 6.07%, however, it was discovered that 72.18% of the BUSCO genes had “duplication” in the A. calamus genome. According to BUSCO analysis, suggest that both Acorus genome assemblies are nearly complete with respect to gene space. Both the A. gramineus and A. calamus assemblies based on the Hi-C data are of high quality. The 12 chromosomes of A. gramineus ranged in size from 13.73 Mb to 32.55 Mb, with a scaffold N50 value of 24.59 Mb. The 22 chromosomes of the A. calamus that was clustered based on the K-mer sequence was successfully clustered into two subgenomes. There were 12 chromosomes in sub-genome B and 10 in sub-genome A. The size of A. calamus sub-genome A is 323.33 Mb and the length of the 10 chromosomes Hi-C assemblies ranges from 21.69 Mb to 45.83 Mb, with a scaffold N50 value of 29.86 Mb. The size of A. calamus sub-genome B is 360.99 Mb and the length of the 12 chromosomes Hi-C assemblies ranges from 24.30 Mb to 33.96 Mb, with a scaffold N50 value of 29.84 Mb.
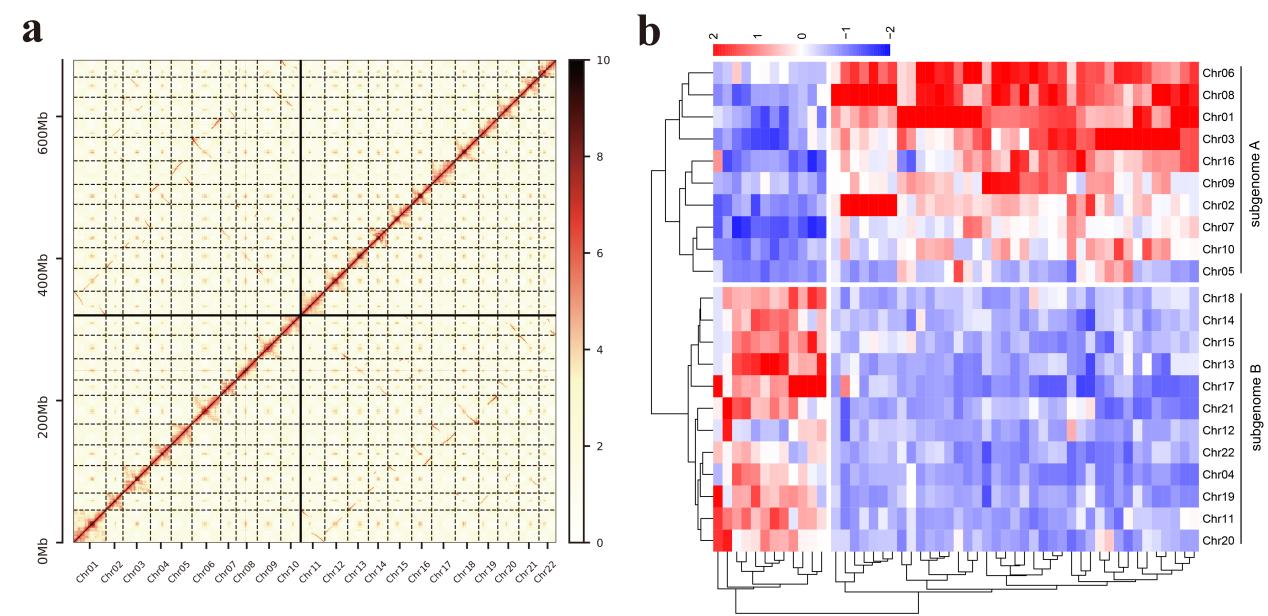
Figure 2. Hi-C scaffolding of the allotetraploid A. calamus genome and sub-genome reconstruction (see the article of Ma et al. for details).
The genome of the A. gramineus contains 25,090 genes, 21,743 genes in A. calamus subgenome A and 24,322 genes in subgenome B. The number of complete BUSCO genes of each A. calamus subgenome is lower than that of the combination of the two A. calamus subgenomes, suggesting that each subgenome has been undergoing reciprocal gene loss after allopolyploidization9-13. The gene ancestor of the Acorus species underwent a whole genome duplication (WGD) in 41.7 Mya, which was not shared by other monocots. The divergence between A. gramineus and the common ancestor of A. calamus A and B occurred approximately 9.9 Mya, while the divergence time for the subgenomes A and B of A. calamus A and A. calamus B, was estimated at approximately 8.5 Mya. Studies revealed that the ancestral chromosome number of A. gramineus and A. calamus was 10. Due to their own WGD, it is suggested that the chromosome number of ancient monocots was 5. Chromosome division events caused A. gramineus and A. calamus B to have 12 chromosomes. A. calamus A and B fused to form an allotetraploid with 22 chromosomes.
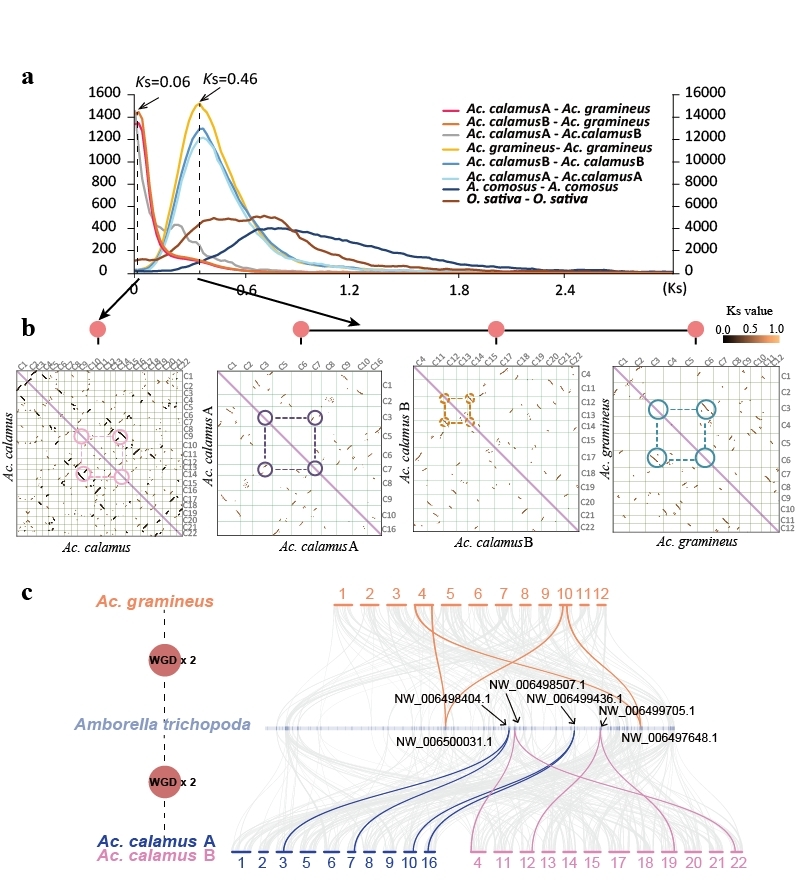
Figure 3. WGD in Acorus (see the article of Ma et al. for details).
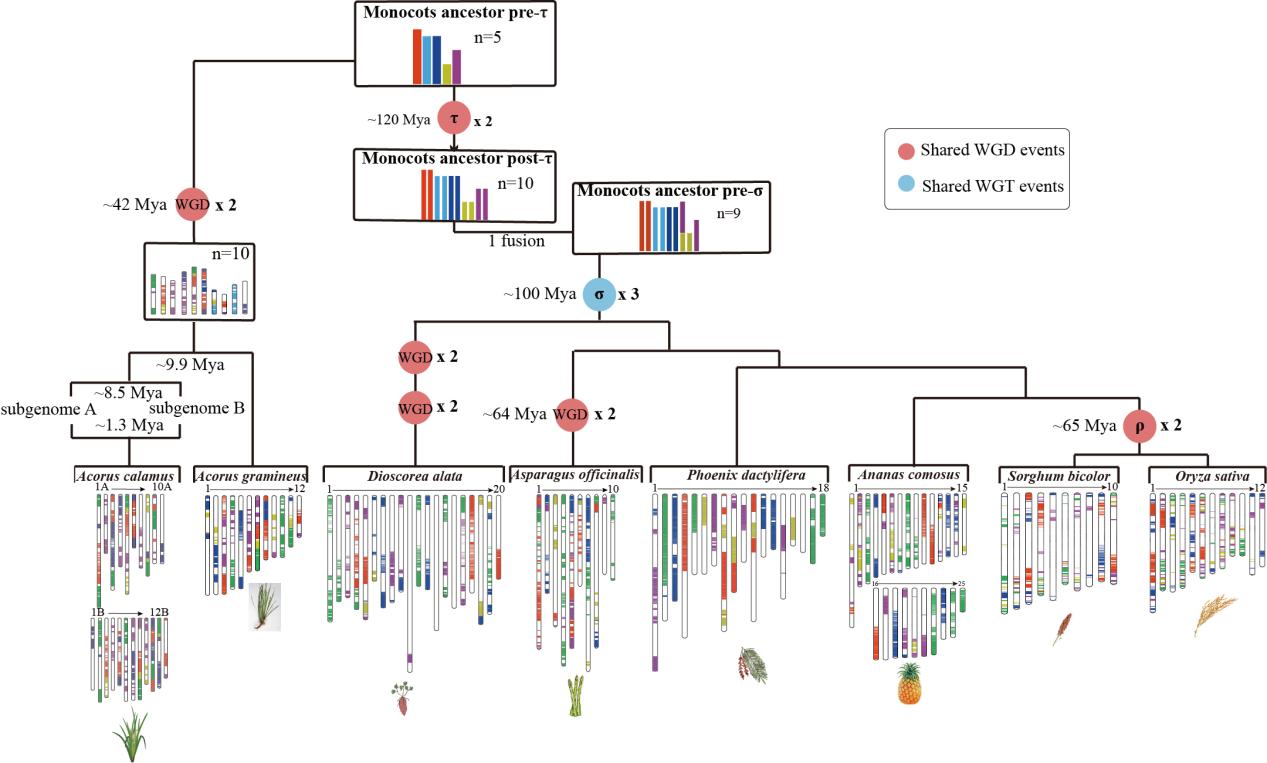
Figure 4. The karyotype evolution in monocots.
This study unveils the occurrence of paleopolyploid events in the evolution of monocots. Include: 1) the τ WGD was shared by most monocots and supposed to have occurred approximately 120 Mya14, 2) the σ WGD was shared by all Poales and supposed to have occurred approximately 110 Mya14-17, 3) the two consecutive WGDs SPα and SPβ in the lineage leading to Sp. Polyrhiza within a short period of time about 95 Mya18,19, 4) the ρ WGD was shared by all Poaceae and supposed to have occurred approximately 70 Mya20, 5) the orchid WGD was shared by all orchids and also supposed to have occurred approximately 74 Mya21, 6) the Acorus WGD, which have occurred approximately 41.7 Mya in the common ancestor of A. gramineus and A. calamus. All the ancient WGDs reported in monocots above are younger than the divergence between Acorales and other monocots, estimated to be at approximately 140 Mya, which agrees with the fact that we could not detect any signal for WGD events older than the Acorus specific ones. For instance, intergenomic collinear analyses between the two Acorus (sub)genomes and the Amborella genome, which has not experienced a WGD since the divergence of angiosperms, only showed two collinear segments in an Acorus (sub)genome to one collinear segment in the Amborella genome, in support of a single WGD duplication shared by Acorus.
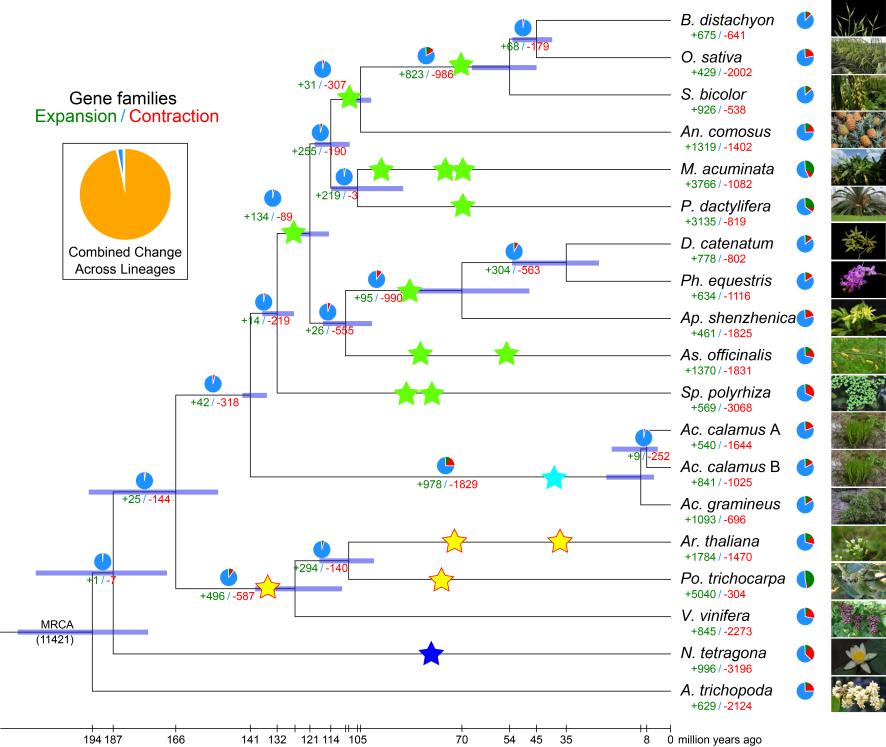
Figure 5. Phylogenetic tree showing divergence times and the evolution of gene family size in 19 species.
By comparing collinearity between A. calamus A, A. calamus B, and A. gramineus, we inferred the karyotype of the most recent common ancestor (MRCA). Considering the minimal collinearity between the A and B subgenomes of A. calamus, and the current A. gramineus genome, it is plausible that the two genome ancestors of A. calamus are not derived from A. calamus itself, but likely from two distinct diploid ancestors, which might have existed in an ancient lineage. The study also collected transposable elements (TEs) from the two subgenomes of A. calamus and assessed their divergence rates22,23, and estimated the time of allopolyploid events, which were hybridization events. It was determined that the two ancestors had differentiated prior to 8.5 Mya, and hybridized around 1.3 Mya to create the current allotetraploid A. calamus. Further analysis showed that subgenome B exhibited fewer gene losses, and after undergoing strong purification selection, it displayed increased gene expression in the promoter region and reduced CG methylation levels. These findings indicated that subgenome B of A. calamus possessed certain advantages, revealing asymmetrical evolution within the allotetraploid A. calamus genome. However, despite sharing homologous genes, these genes have distinct functions, and the two subgenomes exhibit functional complementarity.
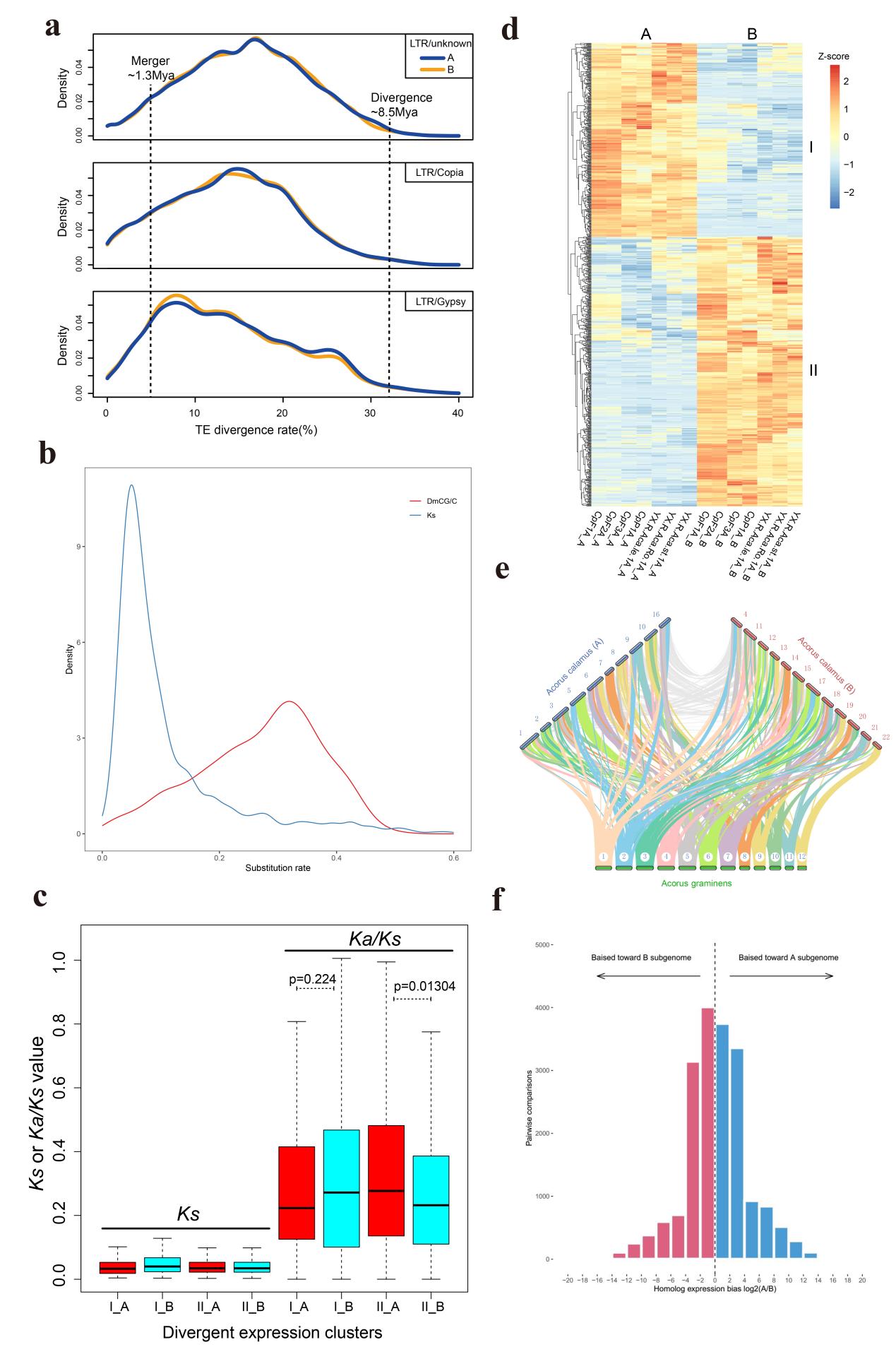
Figure 6. The time of allotetraploidization and DNA methylation in homoeologous expression bias in A. calamus (see the article of Ma et al. for details).
As Acorus is a sister group to the rest of the monocots, its floral organs possess six perianth segments and stamens arranged in two whorls. In total, 90 and 90 putative MADS-box genes were identified in A. gramineus and A. calamus with 45 in subgenome A and 45 in subgenome B, respectively. The expression pattern of the ABCDE gene was like that observed in rice. Class B genes were predominantly expressed in the second and third whorls, class C genes in the third and central whorls, while class E genes were expressed in all flower whorls. These results suggested that MADS-box genes from these subfamilies create the basic blueprint of monocot floral development and form a very interesting system to study the evolution of monocot floral morphogenesis.
Woodiness, a secondary xylem derived from vascular cambium, has been gained and lost multiple times in angiosperms but has been lost in the MRCA of all monocots, such as Nelumbo nucifera, and monocots involved in vascular cambium formation, genes such as OBP1, TMO5, ReVOLUTA, MOL1, and PEAR1 have been lost. Monocots have further lost BEN1 and BDL genes compared to other angiosperms. Eudicots (Arabidopsis and Populus) retained CLE41/44, CEV1, PRR1 and AIL6/7, which are involved in the formation of vascular cambium and secondary cell wall. Similar to N. colorata, monocots have lost genes related to vascular cambium development, which may explain their scattered vascular bundles in the stem.
Angiosperm mesoderm plants are differentiated based on the number of cotyledons: monocots have only one cotyledon, while dicotyledonous plants have two. In dicotyledonous plants, the aerial parts of the seedlings are symmetrical, with two cotyledons positioned between a shoot apical meristem (SAM) 24. The amplification of PIN1 and PID genes, as well as the contraction of CUC gene, specifically affect SAM formation, causing abnormal occurrence sites and resulting in cotyledon formation in monocots 25.
The results show that both Acorus species lost all components in the immune signalling complex except SAG101, which is similar to what has been observed in aquatic monocots and eudicots. The reason that expressions of biosynthetic strigolactone (SL) genes were not highly detected in the roots might be that SLs stimulate early symbiotic responses in both of symbionts but not at the stable stage of symbiosis. These findings uncover the molecular mechanisms behind Acorus and monocots’ ability to adapt to wet environments.
These results showed that early in monocot evolution, species already exhibited many genomic features related to flower development and cotyledon evolution, vascular cambia, secondary xylem development, adaptation to wetland environments, providing fundamental insights into the origin, evolution and diversification of monocots.
This post is jointly written by Yuanyuan Li and Zhong-Jian Liu at Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization at College of Landscape Architecture and Art, Fujian Agriculture and Forestry University.
References
- Givnish, T. J. et al. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot.105, 1888–1910 (2018).
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc.161, 105–121 (2009).
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc.20, 1–20 (2016).
- Cheng, Z. et al. From folk taxonomy to species confirmation of Acorus(Acoraceae): evidences based on phylogenetic and metabolomic analyses. Front. Plant Sci. 11, 965 (2020).
- Plants of World Online: Acorus. (https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:26671#children).
- Wang, H., Li, W. L., Gu, Z. J. & Chen, Y. Y. Cytological study on AcorusL. in Southwestern China, with some cytogoegraphical notes on A. calamus. J. Integr. Plant Biol., 43, 354 (2001).
- Morin, N. R. (Ed.). Flora of North America: North of Mexico Volume 22: Magnoliophyta: Alismatidae, Arecidae, Commelinidae (in Part), and Zingiberidae, pp151 (OUP USA, 1993).
- Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210–3212 (2015).
- Edger, P. P., Pires, J. C. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res.17, 699–717 (2009).
- Jiao, Y., Paterson, A. H. Polyploidy-associated genome modifications during land plant evolution. Philos. Trans. R. Soc. Lond., B, Biol. Sci.369, 20130355 (2014).
- Blanc, G., Wolfe, K. H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691 (2004).
- Aravind, L. et al. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. U.S.A.97, 11319–11324 (2000).
- Xu, P. et al. The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat. Commun.10, 1–11 (2019).
- Ming, R. et al. The pineapple genome and the evolution of CAM photo synthesis.Nat. Genet. 47, 1435–1442 (2015).
- Paterson, A. H. et al. The Sorghum bicolorgenome and the diversification of grasses. Nature 457, 551–556 (2009).
- Tang, H., Bowers, J. E., Wang, X. & Paterson, A. H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage.Proc. Natl. Acad. Sci. USA 107, 472–477 (2010).
- Mckain M. R. et al. A phylogenomic assessment of ancient polyploidy and genome evolution across the Poales. Genome Biol. Evol.8, 1150–1164 (2016).
- Wang, W. et al. The Spirodela polyrhizagenome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 13, 1–13 (2014).
- Michael, T.P. et al.Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 89, 617–635 (2017).
- Wang, X. et al. Genome alignment spanning major Poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant8, 885–98 (2015).
- Zhang, G. Q. et al. The Apostasia genome and the evolution of orchids. Nature 549, 379–383 (2017).
- Ye, C. Y. et al. The genomes of the allohexaploid Echinochloa crus-galliand its progenitors provide insights into polyploidization-driven adaptation. Mol. Plant13, 1298–1310 (2020).
- Jiao, Y., Paterson, A. H. Polyploidy-associated genome modifications during land plant evolution. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 369, 20130355 (2014).
- Furutani, M. PIN-FORMED1and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030 (2004).
- Yang, J., Wang, H., Yan, G., & Qin, Y. Callus induction and differentiation from the cotyledon of Capsicum annuum L. J. Jilin Agric. Uni.22, 51–61 (2000).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in