After the Paper | From Paper to Industrial-scale Platform: a 7-year Behind the Paper Journey from iPANDA to PandaOmics AI-Powered Target Discovery Platform
Published in Bioengineering & Biotechnology

PandaOmics: Behind the Development of Insilico Medicine’s Sophisticated Tool for Turning Complex Biological Data Into Novel Therapeutic Targets
PandaOmics is an artificial intelligence (AI)-driven target discovery platform developed by Insilico Medicine that applies deep learning models to identify therapeutic targets associated with a given disease through a combination of omics data analysis put in the context of prior information coming from publications, clinical trials and grant applications. The algorithm optimizes for the best potential therapeutic targets by scoring results on factors such as: novelty, confidence, commercial tractability, druggability, safety, and other key properties that drive target selection decisions. PandaOmics has been used to identify new targets for cancer, amyotrophic lateral sclerosis (ALS) and COVID-19 and related variants, among other diseases, and the novel target it discovered for idiopathic pulmonary fibrosis has been developed into a lead drug candidate (designed through Insilico Medicine’s Chemistry42 AI platform) that is currently in Phase 1 clinical trials, the first AI-discovered and AI-designed drug to reach this milestone.
Here is the story of how PandaOmics was developed – from its early days as a theoretical algorithm to a validated new means for identifying novel targets to treat some of the most difficult-to-treat diseases, including cancer, fibrosis, and ALS.
iPANDA: The paper that inspired the development of PandaOmics target discovery platform
PandaOmics had its start in 2016, when Insilico Medicine published what was called the Insilico Pathway Activation Network Decomposition Analysis or iPANDA algorithm for biology-justified dimensionality reduction.The algorithm allowed researchers to quickly and efficiently analyze signaling and metabolic pathway perturbation states using gene expression data.
To demonstrate the effectiveness of iPanda, a team from Insilico Medicine worked with collaborators from the Johns Hopkins University, Albert Einstein College of Medicine, Boston University, Novartis, Nestle and BioTime to demonstrate that iPANDA provided significant noise reduction in transcriptomic data and identified highly robust sets of biologically relevant pathway signatures for breast cancer patients according to their sensitivity to neoadjuvant therapy. Breast cancer is the second most common cancer in the U.S. and the second leading cause of cancer death in women. It represents a large unmet need for the development of new highly robust transcriptomic data analysis methods. The results from this research were published in Nature Communications in November 2016.
While others were similarly engaged in using deep learning to tackle complex biological problems, most were employing a piecemeal approach. To create a system that could effectively sort through transcriptomic data and produce biologically meaningful results was a puzzle that required an innovative approach. With iPANDA, Insilico Medicine devised a means for scoring quality metrics within the pathway activation that allowed it to find biologically meaningful targets that met criteria for highly discriminative pathway markers, including biological relevance, cumulative effect, batch effect minimization, consistent performance at the platform- , species-, tissue- and experiment-level, and the ability to reduce the dimensionality of gene expression data for the deep neural networks.
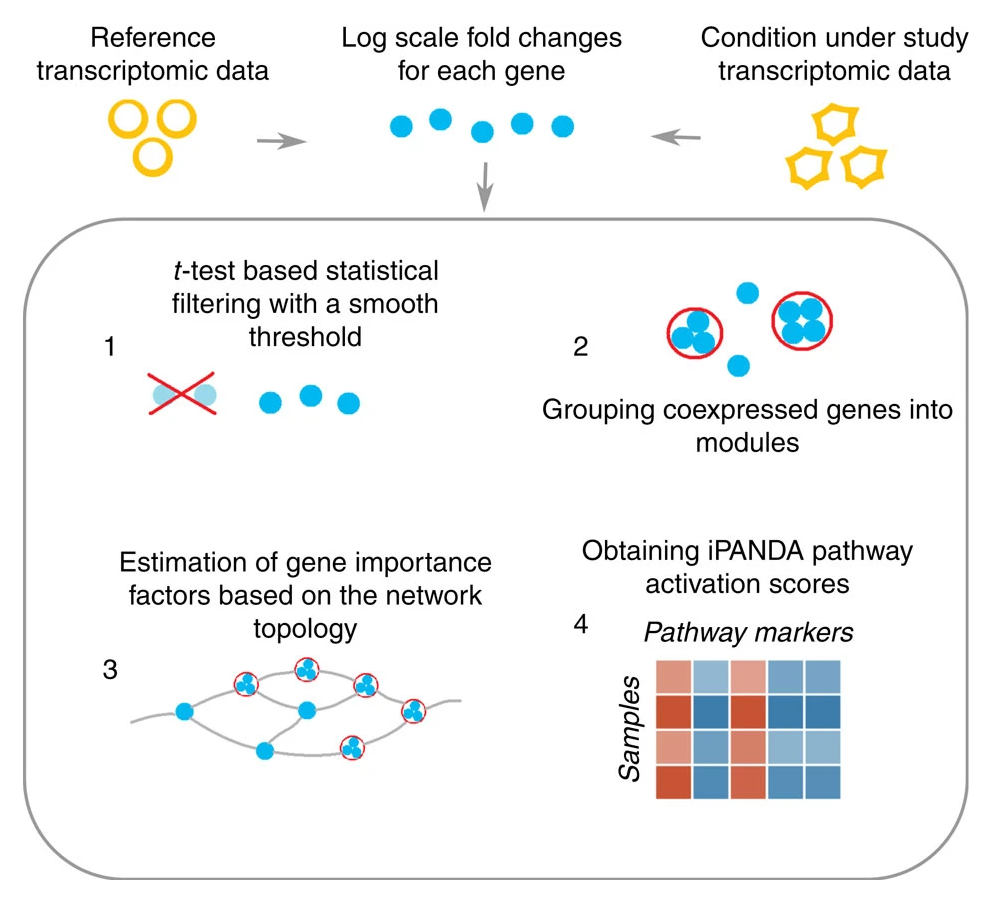
The study showed the effectiveness of iPANDA’s model, which integrated different analytical concepts while also exploiting statistical and topological weights for gene importance estimation. The approach groups genes into modules and then assesses the contribution of gene units (including gene modules and individual genes) to pathway activation by computing their fold changes in logarithmic scale, topological and statistical weights.
Ultimately, Insilico Medicine was able to demonstrate that iPANDA reduced noise while preserving biologically relevant features by performing an analysis of the MAQC data set, which contains data for the same cell samples processed using various transcriptome profiling platforms.
Applied to the breast cancer data, the iPANDA algorithm produced a highly consistent set of pathway markers across the data sets used in the study. In particular, the iPANDA values for 19 and 8 pathways for ERN HER2P and ERN HER2N breast cancer types, respectively, can be utilized as paclitaxel response classifiers with AUC values higher than 0.7 for all data sets examined. All third-party algorithms tested (including GSEA, SPIA, DART, ssGSEA and PLAGE) failed to obtain even a single pathway marker (with the AUC threshold equal to 0.7) common for all data sets examined (for both cancer types), suggesting that the iPANDA algorithm is an advantageous method for biologically relevant pathway marker development compared with other pathway-based approaches.
The evolution of iPANDA into PandaOmics
Once we had established a proven means of identifying relevant biological pathways for diseases, we began to expand the system’s predictive capabilities through additional data–both from existing sources and from our own experiments.
We saw that the iPANDA method of transcriptomic data analysis on the signaling pathway level was not only useful for discrimination between various biological or clinical conditions, but could also aid in identifying functional categories or pathways that may be relevant as possible therapeutic targets.
From 2016 until PandaOmics was officially launched in 2020, Insilico Medicine published broadly in leading scientific journals as it further expanded its target discovery capabilities. A paper published in 2016 by the American Chemical Society showed that Insilico’s deep learning applications could be used to predict the pharmacological properties of drugs across different biological systems and conditions using transcriptomic data.
In 2018, the Insilico team used iPANDA to identify bifunctional immune checkpoint-targeted antibody-ligand traps for cancer immunotherapy – results were published in Nature Communications. In 2019, they published research in Springer showing how the algorithm could discern aberrantly activated signaling events in gallbladder cancer and found that PIM1 could be a therapeutic target.
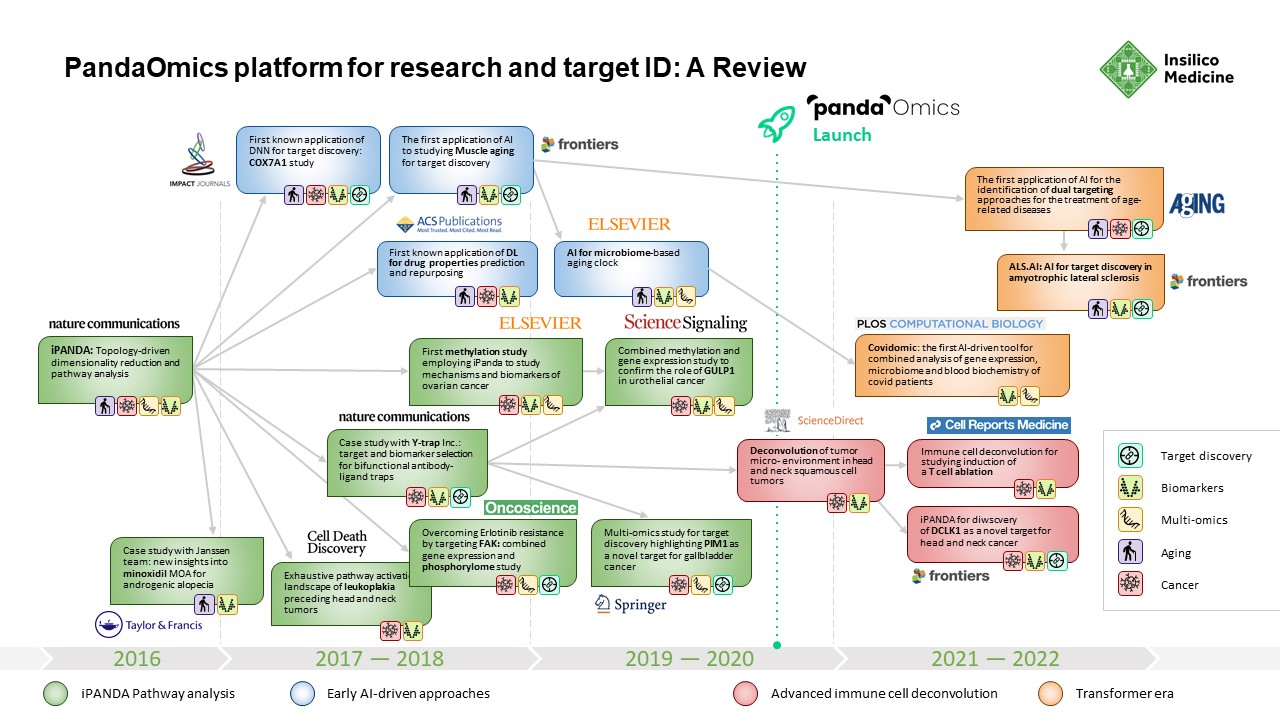
In 2020, PandaOmics was officially launched, drawing on trillions of data points, built up over the intervening years. This includes 5 million omics data samples, including transcriptomics, genomics, epigenomics, proteomics, and single cell data generated by the scientific community; 1.3 million compounds and biologics; 3.8 million patents; over $1 trillion in grants; 342,000 clinical trials; and over 30 million publications.
PandaOmics begins with iPANDA’s proprietary pathway analysis to infer pathway activation or inhibition. Then, its AI algorithm analyzes all data and significant genes from an experiment in the context of the disease being studied and identifies a number of actionable drug target hypotheses. The system then evaluates the targets using molecular and text evidence and predicts the likelihood of that target entering Phase 1 clinical trials within the next five years. PandaOmics also predicts the “hotness” of a particular target based on the attention it is receiving in the scientific community.
In 2021, Insilico researchers performed a comprehensive transcriptome-based computational analysis on data from hundreds of patients with head and neck squamous cell carcinoma, and discovered that high DLCK1 expression positively correlates with NOTCH signaling pathway activation and could serve as a potential therapeutic target. The study was published in Frontiers in Oncology.
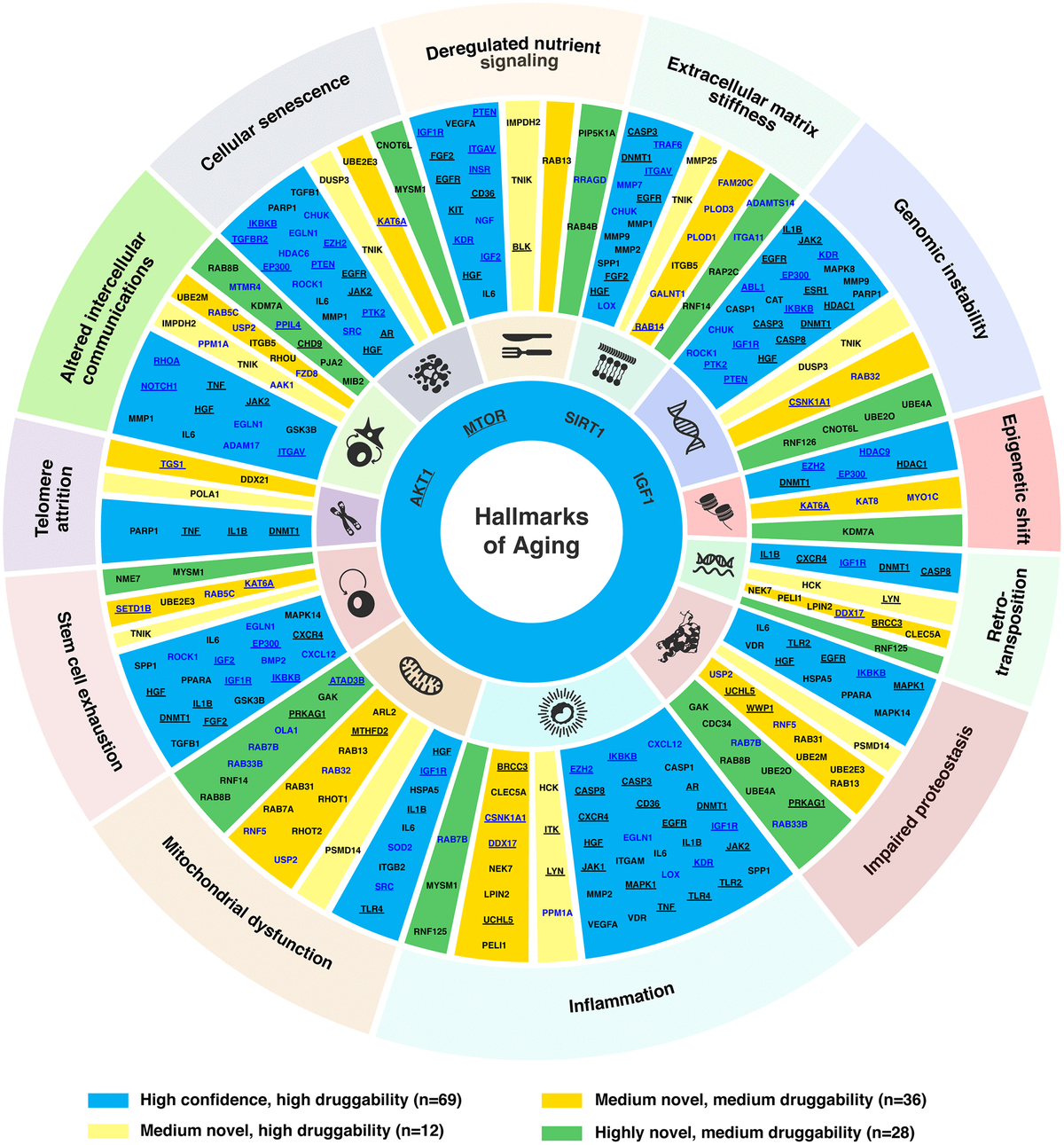
In just two months’ time, Insilico also used PandaOmics to uncover multiple targets specific to both aging and disease, a finding that could lead to the development of dual-purpose therapeutics. Identifying such targets could provide added effectiveness for older patients who are suffering from disease, as well as delaying or reversing certain aging symptoms in addition to treating disease symptoms. In the study, Insilico deployed PandaOmics to perform target identification for 14 age-associated diseases and 19 non-age-associated diseases across multiple disease areas to identify targets of age-associated diseases. PandaOmics revealed 145 genes that were considered potential aging-related targets and mapped these into corresponding aging hallmarks, including 69 high confidence targets with high druggability, 48 medium novel targets with high or medium druggability, and 28 highly novel targets with medium druggability. The results were published in March 2022 in the journal Aging.
The AI-powered platform for the pharma companies and academic researchers to accelerate their programs
Since it officially launched in 2020, PandaOmics has been employed by numerous top pharma companies and by academic researchers to identify novel targets for their programs.
Pfizer entered into a research collaboration with Insilico in January 2020 to explore new targets for diseases with high unmet needs using PandaOmics. In August 2021, Insilico announced a research partnership with Arvinas to use PandaOmics to discover novel PROTACs. In January 2022, a collaboration agreement was announced between Insilico and Fosun Pharma to use PandaOmics to discover four biological targets and to co-develop a QPCTL program at Insilico.
According to the collaboration agreement, Insilico will deliver a preclinical candidate for the QPCTL program and advance it to the IND stage, after which Fosun Pharma will conduct human clinical studies and co-develop the candidate globally. In parallel, Fosun Pharma's R&D team will nominate four therapeutic targets to be assessed by Insilico's AI platform and R&D team, who are responsible for advancing these drug candidates to IND stage.
Insilico’s PandaOmics AI target discovery platform was also recently used by a consortium of top ALS researchers from Harvard, Johns Hopkins, Mayo Clinic, University of Chicago, Shanghai University and other institutions, in order to find new targets for the debilitating neuromuscular disease. The platform drew on comprehensive patient data collected by the research group Answer ALS. The AI software found 17 high-confidence and 11 novel therapeutic targets. The results were published in June 2022 in the journal Frontiers in Aging Neuroscience.
Advancing the first AI-discovered and AI-designed drug to human trials
Among the platform’s most important advances in end-to-end AI drug discovery has been the identification of Insilico Medicine’s lead drug candidate for treating idiopathic pulmonary fibrosis. In just 18 months, PandaOmics discovered a novel pan-fibrotic target and Insilico’s AI drug design platform Chemistry42 designed a novel corresponding drug candidate for 1/10 of the typical cost of such drug development. PandaOmics sorted through millions of data files, including patents, research publications, grants, and databases of clinical trials, and revealed 20 potential targets for IPF that researchers narrowed down to one novel intracellular target and prioritized for further analysis.
The lead drug candidate has since been through extensive target validation, and is currently in Phase 1 trials in New Zealand and China, the first AI-discovered and AI-designed drug to reach this milestone.
Paper Reference:
Ozerov, I., Lezhnina, K., Izumchenko, E. et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nat Commun 7, 13427 (2016). https://doi.org/10.1038/ncomms13427
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in