After the Paper | Looking back on co-abundance binning and what it meant for microbiomics and its authors
Published in Bioengineering & Biotechnology
We discovered co-abundance species binning already in 2010 when we, in the MetaHIT consortium, received the first large set of shotgun sequenced metagenomic samples, which was used to reconstruct the first human intestinal gene catalogue (Qin et al., Nature, 2010). Coming from the microarray field it seemed as the most natural thing to cluster genes by abundances. Still, it was a bit of a revelation to see how well it worked for identifying species we knew from reference genomes as well as novel species. Most of the data did not map to anything known and the abundances for genes from reference genomes were all over the place. Which is not very satisfying if you wanted to profile the microbiome composition. So we turned the logic on its head, looking for a signal that was co-abundant, and voila, there was the metagenomic species (MGS).
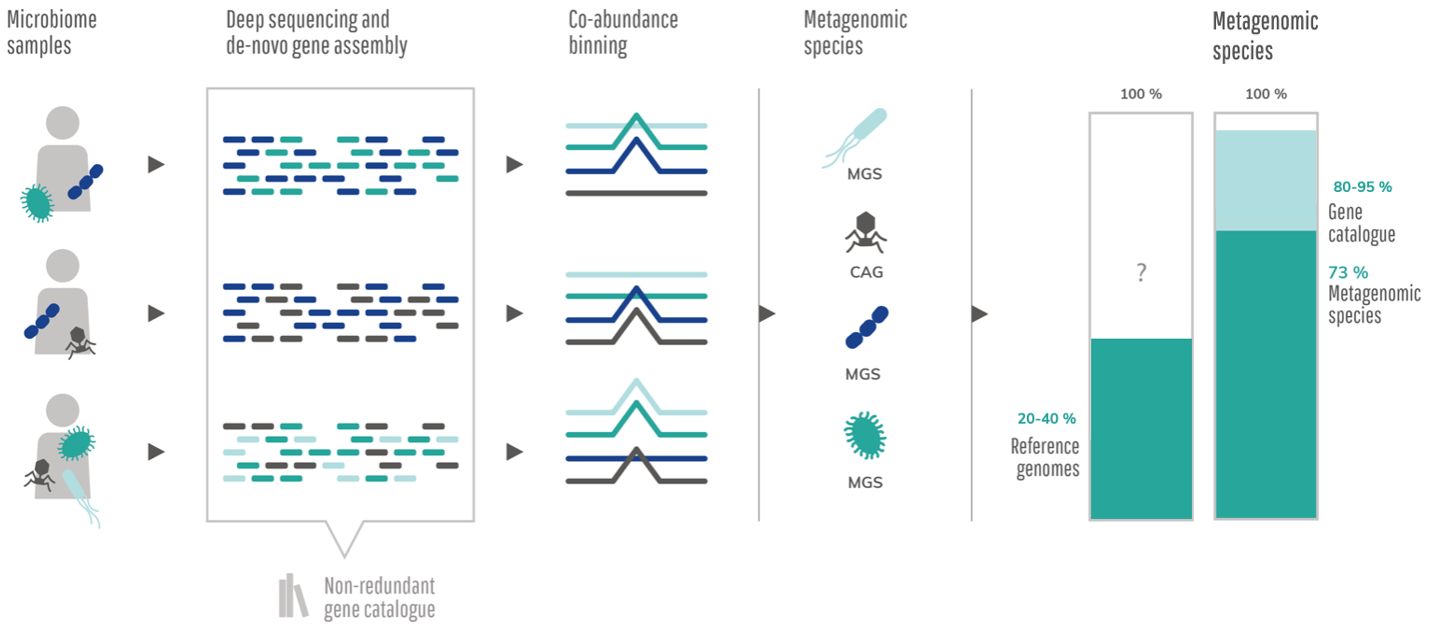
Figure 1 : Co-abundance based clustering used to decipher the known and unknown intestinal microbial species (Nielsen et al., Nature Biotechnology, 2014). The metagenomics species and gene catalog describes a significantly higher proportion of the metagenomics reads than reference based approaches.
It took three years before the method finally got published in 2014. More medical focused stories were simply prioritized. Which was perhaps reasonable, but frustrating for us. Luckily it came out in the end. Right after it came out in Nature Biotechnology we presented the story at different conferences. At that time the idea of co-abundance binning across a large complex microbiome dataset was revolutionary, but it was received with much excitement. Suddenly, we could explain most of the microbiome data and observe all of these novel species, many of which have since been cultivated and named.
The paper inspired many others to create similar methods for binning metagenomics data, including an abundance-based binning method for microbial pangenome reconstruction (Plaza Oñate et al., Bioinformatics, 2019), a deep learning method published this year (Nissen et al., Nature Biotechnology, 2021), and more importantly it led to many biomedical discoveries. The focus of most current metagenomics binners differ somewhat from our original focus. Although the paper presented 238 high-quality microbial genomes, which was the largest batch deposit to EBI by then, it was compositional profiling of the microbiome that was in focus. Focusing on empirically well behaved genes, i.e. genes that were highly co-abundant we mostly found the core genomes of the MGS. Most of the original MGS comprised about 90% of the genes of corresponding reference genomes. The pangenome is simply not very good for abundance profiling. It was better to split these elements out in smaller co-abundance gene groups or CAGs.

Figure 2 : The within species phylogeny of Akkermansia muciniphila calculated from single nucleotide variations (SNVs) of its signature genes shows that the optional gene set of the species correlates with the phylogeny. The figure was provided by Pernille Neve Myers, Clinical Microbiomics.
Many of these corresponded to phages whereof many were later confirmed, but others were simply the pangenome of other MGS. Today, one would probably consider a phylogenetic model for this level of variation.
For us personally, the paper gave us the chance to speak at a series of scientific conferences, on national TV, and in countless news outlets, and to be involved in a number of other research projects.
For H. Bjørn Nielsen this ultimately resulted in his current position as CSO in Clinical-Microbiomics. Clinical-Microbiomics provides microbiome research as a service, and the co-abundance idea of the 2014 paper is still a cornerstone in our profiling of the microbiome. We have improved the method: included the entire gene set of the MGS and uses SNV calling and phylogenetic models to resolve sample specific populations, or strains as they are commonly called. But the idea of using a signature gene set for accurate abundance profiling is still a key element.
For Mathieu Almeida, the publication fueled an academic career. In 2013, after defending his phD under the supervision of Dr. Pierre Renault and Dr. S. Dusko Ehrlich at INRAE (France), he joined Dr. Mihai Pop’s team at the University of Maryland CBCB institute (USA). There, he focused on providing genomic context for the human microbiota « most-wanted » microorganisms – uncultured organisms that are common in the healthy microbiota (Almeida et al., ISME, 2016). Today, Mathieu Almeida is a Research Fellow at INRAE MetaGenoPolis, in charge of the French Gut Project bioinformatic analysis (http://mgps.eu/projects/), aiming at collecting and exploring 100,000 French human gut microbial metagenome before 2025, to improve human health prognosis and the definition of an healthy microbiota.
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in