All for one and one for all, united we stand, divided we fall: How does a cell express only one gene out of a large gene family?
Published in Microbiology, Cell & Molecular Biology, and Genetics & Genomics

In our latest work, we did a deep-dive into characterisation of an inter-chromosomal bridge comprising two sub-nuclear foci-forming proteins that sustain singular antigen expression in African trypanosomes.
Single gene choice underpins our very own sense of smell or the specificity of our immune response. Strikingly, this mechanism is also exploited by many pathogens to undergo antigenic variation, one of the most sophisticated virulence mechanisms in infection biology. Some of the deadliest parasites, such as African trypanosomes and malaria parasites, can systematically alter the identity of proteins displayed to the host immune system. There are two key aspects for successful antigenic variation: 1) the mutually exclusive expression of one antigen isoform from myriad possibilities and 2) the periodic switching of expression of one antigen isoform to another.
African trypanosomes are masters of disguise. They express their variant surface glycoprotein (VSG), forming a dense surface coat, in a monogenic fashion from a repertoire of >2,000 genes encoding VSG isoforms. Strikingly, a high rate of transcription by RNA-polymerase-I and post-transcriptional regulation ensure that the active VSG gene produces the most abundant protein of the trypanosome cell (10 million molecules on the surface of each individual cell, amounting to 10% of total protein).
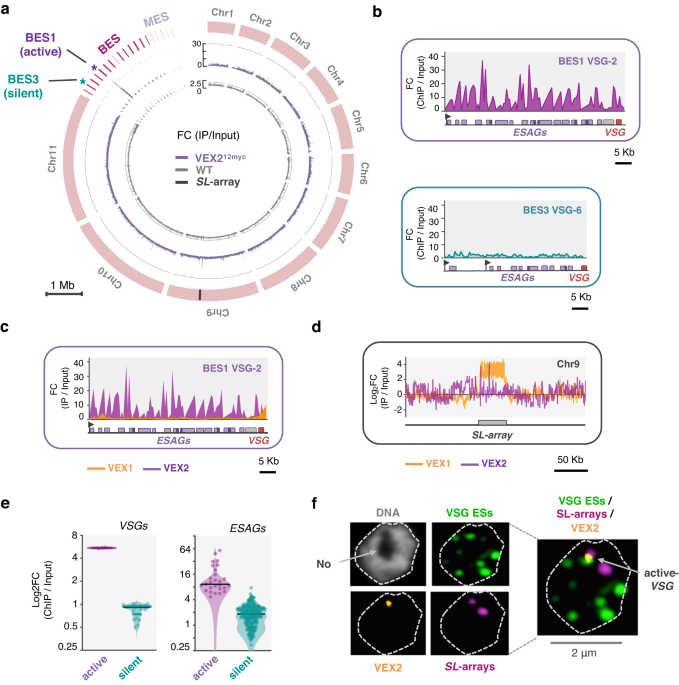
Previously we showed that physically bringing the single active antigen-coding-locus in proximity to an RNA maturation ‘hotspot’, only one antigen variant is expressed at any given time and at very high levels through post-transcriptional enhancement – in simple terms, trypanosomes assemble a sub-nuclear super-factory for antigen production (PMID: 33432154). Notably, two players stood out in this ‘nuclear enterprise’: VSG exclusion factors 1 and 2 (VEX1 / VEX2) form discrete protein foci that colocalise with the RNA trans-splicing and VSG transcription compartments, respectively, suggesting that they might bridge and tether the two DNA loci. In the current study, we used multiple different experimental approaches, including protein affinity purification followed by mass spectrometry analysis, ChIP-Seq, single cell RNA-Seq, super resolution microscopy (3D-SIM) and biochemical assays (including native gels, size exclusion chromatography, protein turnover assays) to tackle this question.
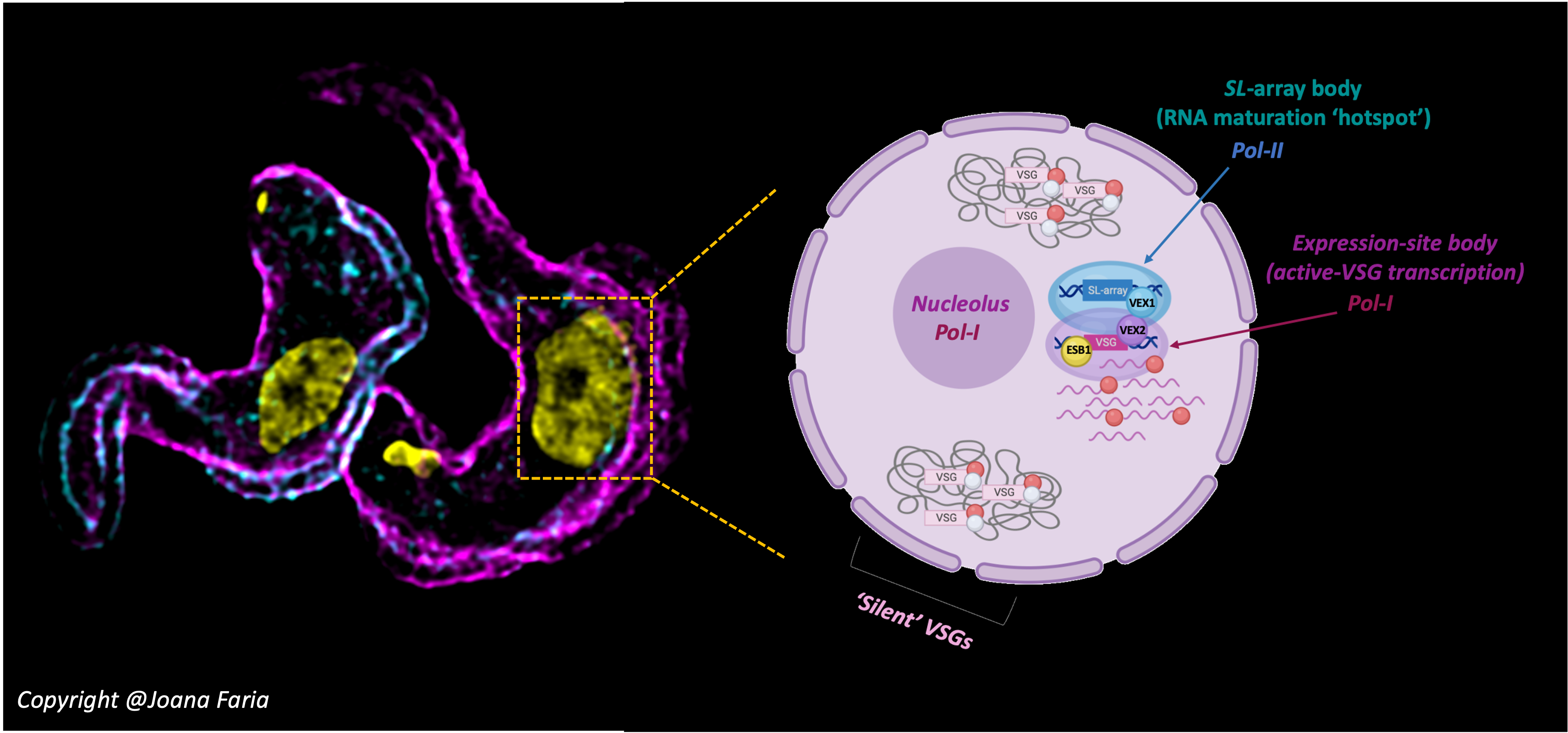
We show in this work that VEX2 is a VSG allele-selective and self-limiting factor that, together with VEX1, forms a physical bipartite inter-chromosomal bridge to support single gene choice. Further, we show that VEX1 and VEX2 association is dynamic and cell cycle dependent. We propose a model whereby VEX1 selectively stabilises VEX2 in the context of an active VSG transcription-unit, thus limiting the pool of VEX2 available to accumulate at other VSG transcription-units and enforcing monogenic antigen expression. It is also worth noting here that our observations are rather unique, as other proteins have been implicated in chromosome looping, bridging and nuclear organisation, such as CTCF in metazoa, but none of these proteins is thought to be allele selective.
Unsurprisingly, VEX2 depletion results in de-repression of otherwise silent VSGs, as we previously reported (PMID: 31289266). One of the limitations we faced at that time was the inability to assess VSG expression at single cell resolution, therefore potentially missing additional layers of complexity and regulation. In the current study, we used single cell transcriptomics to revisit this question, which revealed intriguing and unexpected insights into the allelic exclusion system. Firstly, we found that the most common phenotype was less promiscuous with simultaneous expression of three to six VSGs per cell (instead of the 15 we can detect in the population), likely limited by the availability of transcriptional and/or RNA processing machineries. Most notably, we found that VSG derepression following VEX2 depletion was more pronounced for those transcription units with two Pol-I promoters, and followed a well-defined hierarchy, showing that VSG promoters drive the competition for activation.
From a personal perspective, this study is very meaningful to me for two very important reasons. Firstly, the use of so many different orthogonal approaches, which was only possible through collaboration with an interdisciplinary team of colleagues, allowing us to approach a complex problem from different angles. Mark and Martin brought in expertise in cryomilling-affinity enrichment proteomics, which they have implemented and optimised in trypanosomes. Haru contributed with his ample expertise in the development of methods for sub-cellular fractionation (particularly nucleolar fractionation), for proteomics analysis. Cat made a significant contribution in the dissection of the VEX1 / VEX2 interaction. Michele provided invaluable help with bioinformatic analyses, especially with the single cell transcriptomics, which presented us with VSG-specific issues that were challenging to circumvent. Secondly, this work marks my transition to independence, as I started the work as a postdoc at the University of Dundee in David’s lab and finished it at the University of York while setting up my own lab – a period I will always look back at with a smile.

So why study singular antigen expression in trypanosomes? Well, it is a fine example of extreme biology. Indeed, such high levels of VSG expression render trypanosomes a highly amenable system to study fundamental mechanisms by which cells selectively activate genes and enhance their expression. We strongly believe that there is plenty of novel biology yet to be unravelled in early branching, differently evolved eukaryotes - hopefully our work will strengthen this view. If you want to learn more, you can read it in full here.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in