Antibiotic Design Inspired by Nature
Published in Microbiology

‘There’s more than one way to skin a cat’. So the proverb goes, suggesting the same end can be achieved in different ways. In our paper, we describe a fascinating example of this happening in nature where, instead of looking to skin a cat, the goal is to compete successfully in bacterial internecine warfare. Specifically, we have discovered how two completely different types of bacteria, one Gram-positive, the other Gram-negative, have figured out a way of making two very different antibiotics with which to fend off bacterial neighbours in precisely the same way. This is an exquisite example of molecular convergent evolution. While the two antibiotics are chemically distinct, one is a cyclic depsipeptide (globomycin), the other a macrocyclic lactone (myxovirescin), remarkably they achieve the very same end of shutting down synthesis of key lipoprotein components of the cell envelope, thereby killing or weakening the bacteria, through promiscuous chemical complementarity.
Whilst it is important scientifically to understand how nature crafts and moulds at the molecular level, as illustrated in this work, our findings have the added benefit of providing drug designers chemical blueprints - pharmacaphores - which are known to work for bacteria in the real world. They can now be used to guide pharmaceutical chemists design new, more effective drugs urgently needed in light of the accelerating global threat of antimicrobial resistance.
Much effort has recently been devoted to tackling ‘undruggable’ targets many of which lack defined binding pockets. The signal peptidase II that takes centre stage in our work, likewise has an open (substrate) binding surface. In its case, nature has figured out at least two ways of targeting it with very high affinity using the cyclic natural products, globomycin and myxovirescin. And they do so in ways that are, at once, similar and distinct.
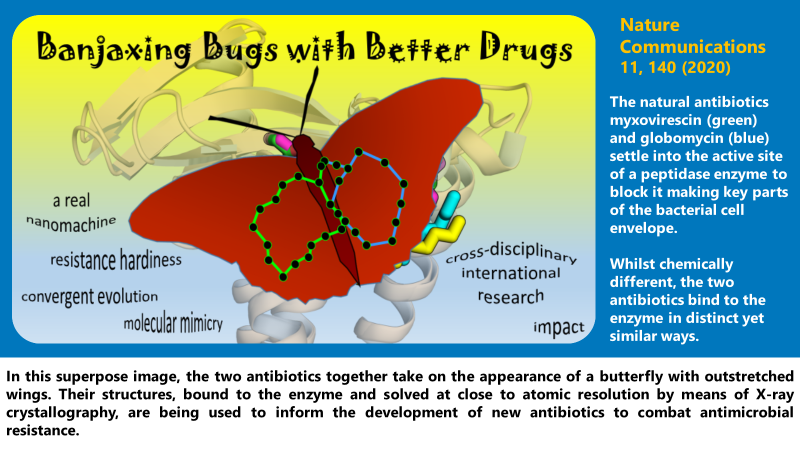 The confluence of ideas to emerge from this study is the product of a cross-disciplinary international research effort involving structural biologists, biochemists, chemists and microbiologists from across the globe. It is based on the following publication:
The confluence of ideas to emerge from this study is the product of a cross-disciplinary international research effort involving structural biologists, biochemists, chemists and microbiologists from across the globe. It is based on the following publication:
Olatunji, S., Yu, X., Bailey, J., Huang, C-Y., Zapotoczna, M., Bowen, K., Remškar, M., Müller, R., Scanlan, E. M., Geoghegan, J. A., Olieric, V. & Caffrey, M. Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin. Nature Communications 11, 140 (2020) https://doi.org/10.1038/s41467-019-13724-y
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in