Antibiotics kill bacteria… by making them lose half of their body
Published in Microbiology
Antibiotics were a scientific breakthrough in the 20th century, and antibiotic resistance is one of the most pressing problems in microbiology in the 21st century. A better understanding of how antibiotics kill cells could inform next-generation approaches to developing better antimicrobial treatments, in addition to helping us better understand prokaryotic cell biology. How antibiotics induce bacterial cell death downstream of binding to the primary drug target may be clearer for some antibiotics than others1. For instance, β-lactams bind to penicillin-binding proteins, which inhibits normal peptidoglycan cell wall synthesis and results in the physical explosion of a bacterial cell2,3. However, other antibiotics, like aminoglycosides and fluoroquinolones, inhibit cellular processes including translation and transcription, and we do not know how exactly inhibition of these processes leads to cell death.
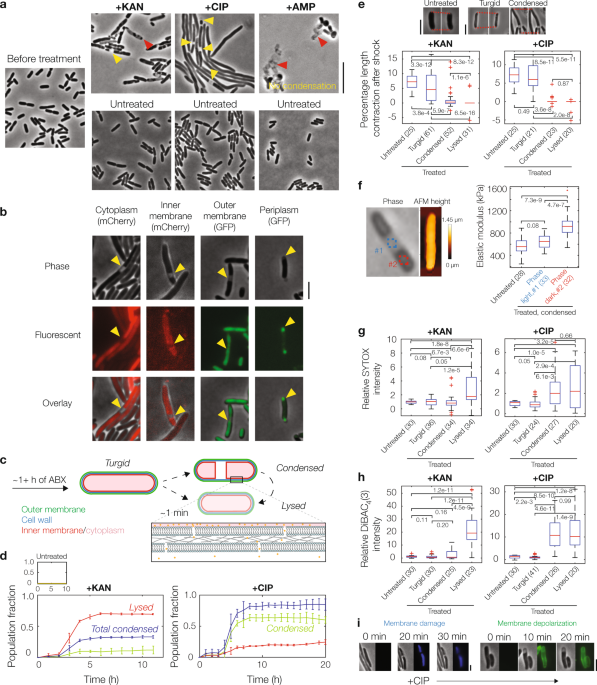
Our study suggests a possible answer. Using long-term microfluidic imaging of aminoglycoside- or fluoroquinolone-treated Escherichia coli cells, we found an intriguing phenotype that could provide us with clues as to how cell death occurs. We observed, in particular, that these bactericidal antibiotics induce cytoplasmic condensation and subsequent lysis in single cells: after several hours of treatment, cells suddenly lose a fraction of their cytoplasm, as manifested by discrete cellular regions becoming phase-light under phase-contrast microscopy. Additional experiments confirm that this condensation is associated with cell envelope shrinkage and possible compactification of cytoplasmic contents, and further suggest that it arises as a result of membrane damage. A biophysical model of cell envelope mechanics and solute outflow confirms that membrane damage can give rise to cytoplasmic condensation. Additionally, the model suggests that only minor damage to the bacterial cell membranes (a loss of 1 in 106 phosholipids) is sufficient to explain the timescale and size of the effect.
Importantly, the phenotype of cytoplasmic condensation is associated with killing. Cytoplasmic condensation is seen in nearly all fluoroquinolone-treated cells, and a fraction (~30%) of aminoglycoside-treated cells, after times corresponding to ≥1 log of killing; we observed more phenotypic variability in aminoglycoside-treated cells ostensibly due to faster lysis and additional ionic interactions of aminoglycosides with the outer membrane, and cannot rule out that more aminoglycoside-treated cells exhibit cytoplasmic condensation than tracked in our protocol. In both cases, however, antibiotic-susceptible cells stop elongating before or once they condense, and cells lyse shortly afterward.
What causes the membrane damage leading to this condensation? A well-known hypothesis in the field is that reactive metabolic byproducts, including but not limited to reactive oxygen species (ROS), are produced as a result of increased cellular metabolism after antibiotic treatment, and these molecules interact promiscuously with cellular components including nucleic acids, lipids, and proteins to contribute to cell death4,5. Consistent with this hypothesis, we found evidence that membrane damage is associated with the accumulation of reactive metabolic byproducts and lipid peroxidation. Lipid peroxidation induced by free radicals may damage the bacterial cell membranes to the extent that appreciable solute outflow and cytoplasmic condensation occur. Once the membranes are damaged in this way, cells cannot recover and are effectively dead. Thus, these findings indicate a sequence of cellular events through which reactive metabolic byproducts lead to physical disruption of the cell envelope and antibiotic-induced cell death.
For details of these experiments, as well as additional microbiological findings, the paper in Nature Communications is here: https://www.nature.com/articles/s41467-021-22485-6
- Kohanski, M. A., Dwyer, D. J. & Collins, J. J. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423-435 (2010).
- Yao, Z., Kahne, D. & Kishony, R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol. Cell 48, 705-712 (2012).
- Wong, F. & Amir, A. Mechanics and dynamics of bacterial cell lysis. Biophys. J. 116, 2378-2389 (2019).
- Kohanski, M. A., Dwyer, D. J., Hayete, B., Lawrence, C. A. & Collins, J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797-810 (2007).
- Stokes, J. M., Lopatkin, A. J., Lobritz, M. A. & Collins, J. J. Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251-259 (2019)
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in