Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants
Published in Microbiology
Since the onset of the COVID-19 pandemic, the spike (S) protein of SARS-CoV-2 has undergone significant antigenic drift, leading to the emergence of multiple variants. In late 2021, the Omicron BA.1 variant emerged, carrying 30 mutations in its S-protein, which enabled substantial immune evasion. Several recurrent mutations, including K417N, L452R, T478K, E484A, and N501, observed in earlier variants, have become fixed in the Omicron S-protein. These mutations are located in the epitopes recognized by several widely induced public antibodies, implying that the antigenic drift of S-protein is closely related to immune pressure. Notably, the mutations S371L/F, S373P, and S375F, which first appeared in the Omicron BA.1 S-protein, are critical for the Omicron nasal cell tropism, S-protein function, and antigen presentation. However, the mechanism driving the emergence of these mutations remain unclear. This study identifies that VL6-57-utilizing antibodies are widely present in the population and target a converge epitope defined by residues S371, S373, and S375, suggesting that this class of public antibodies exerts immune pressure that promotes the introduction of S371L/F-S373P-S375F in Omicron variants.
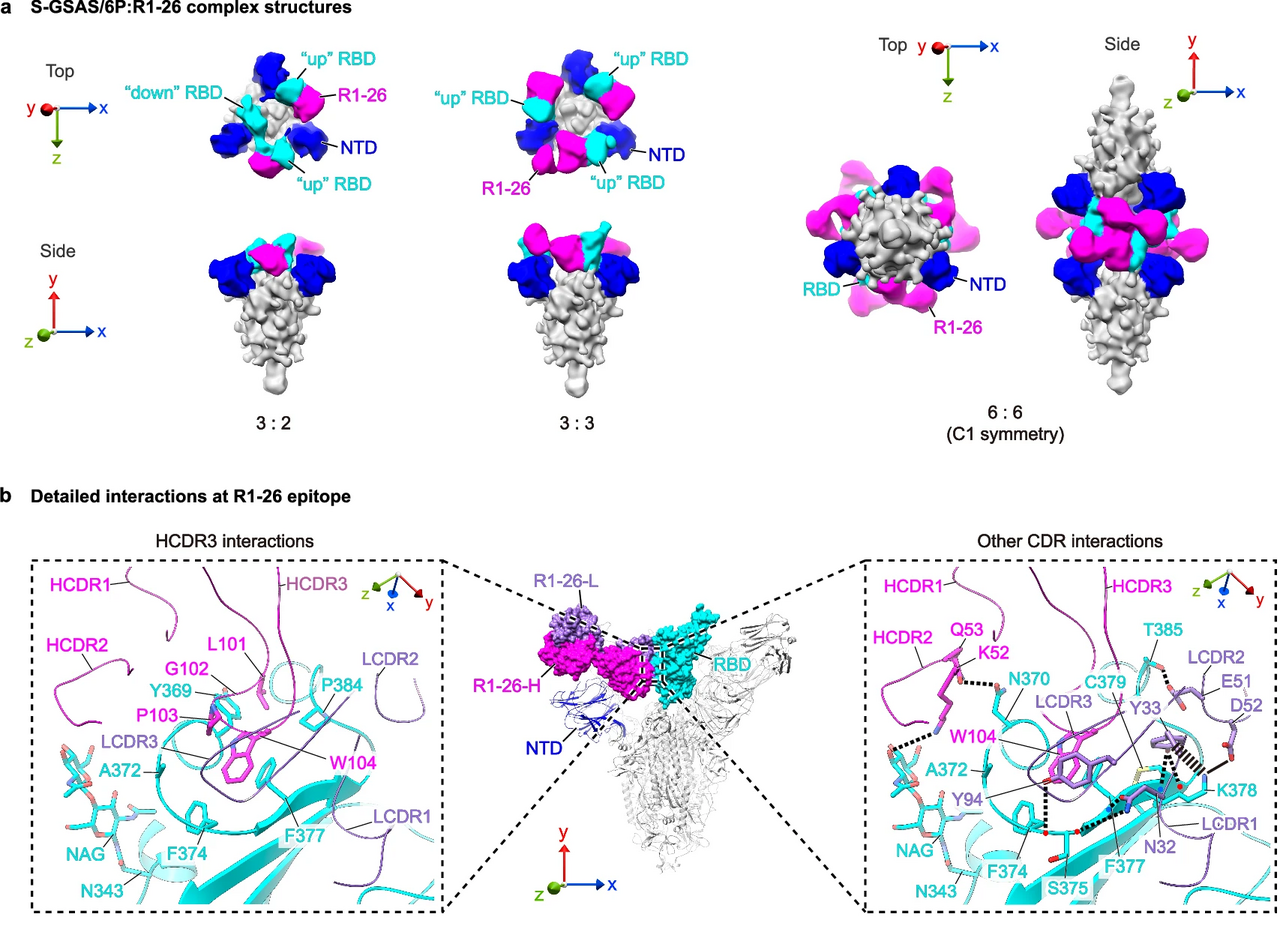
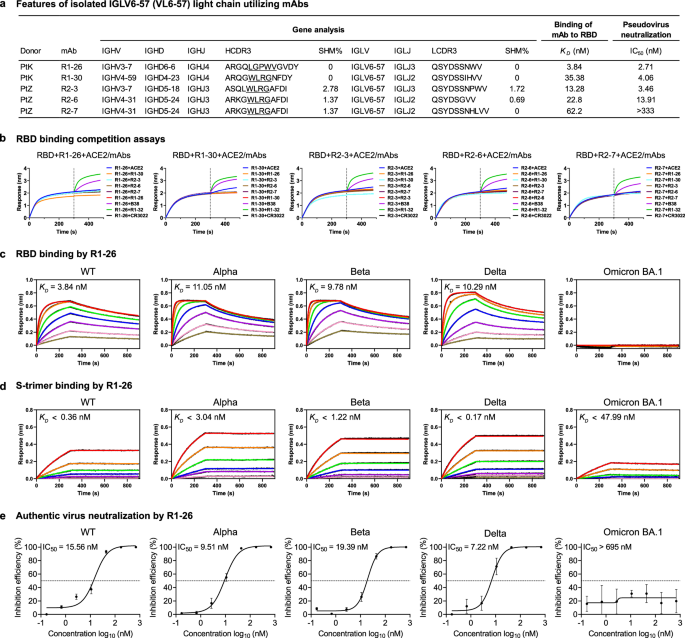
Previously, we isolated 6 RBD-targeting antibodies, R1-26, R1-30, R1-32, R2-3, R2-6, and R2-7 from SARS-CoV-2 wild-type infected patients using phage display. With the exception of R1-32, the other 5 antibodies utilize light chains encoded by VL6-57. Despite variations in their heavy chains, these VL6-57 antibodies share similar hydrophobic motifs in their HCDR3 region, with R1-26 containing “LGPWV” and the others featuring “WLRG”. All 5 antibodies compete with ACE2 and each other for binding to the RBD, suggesting they target overlapping epitopes. Functional studies demonstrated that R1-26 effectively binds to the RBD and S-protein of SARS-CoV-2 variants prior to Omicron BA.1, exhibiting strong neutralization activity against corresponding authentic viruses. Structural analysis revealed that the HCDRs and LCDRs of R1-26 bury comparable surface areas on the RBD, with the hydrophobic HCDR3 and hydrophilic LCDR1 primarily mediating the interaction. Detailed epitope mapping confirmed that R1-26 targets a cryptic epitope on the S-trimer, classifying it as a class 4 antibody. Additionally, R1-26 induces a conformational change of S-protein and inhibits S-protein-ACE2 mediated cell-cell fusion, thereby contributing to its neutralization activity.
To understand antigen binding by VL6-57 light chain antibodies, we surveyed the Protein Data Bank and found that among 376 SARS-CoV-2 S-specific antibody structures, 12 utilize VL6-57 light chains, with 10 belonging to class 4 antibodies. Germline usage analysis shows that VL6-57 is the most frequently used gene in class 4 antibodies, highlighting its importance in antigen binding. BSA analysis reveals that VL6-57 class 4 antibodies derive comparable contributions from both heavy and light chains in epitope recognition, with HCDR3 and LCDR1 regions playing pivotal roles. Structural comparisons indicate that VL6-57 antibodies target convergent epitopes, primarily through shared hydrophobic interactions mediated by HCDR3, and germline motifs in LCDR1 also enhance RBD binding. These results suggest that VL6-57 light chains provide a robust framework for generating class 4 SARS-CoV-2 S-specific antibodies.
An analysis of the Coronavirus Antibody Database identified 290 antibodies utilizing VL6-57 light chains, with 34% featuring 12-AA long HCDR3s and 9-10-AA LCDR3s. Sequence analysis showed significant enrichment of “WLRG” motif within the 12-AA long HCDR3 and “QSYDSS” motif within LCDR3 of these VL6-57 antibodies. Although the LCDR3 covers smaller areas compared to LCDR1, it plays an auxiliary role in supporting HCDR3-mediated antigen binding. Further analysis of immunoglobulin heavy chain (IgH) repertoires revealed the presence of “WLRG” motif-containing IgH sequences in both COVID-19 convalescents and naïve donors, with marked enrichment in COVID-19 convalescents. After SARS-CoV-2 exposure, clonal expansion and class switching of “WLRG” motif-containing IgH sequences were observed. In contrast, the “QSYDSS” motif containing VL6-57 light chains were found at similar levels in both COVID-19 convalescents and naïve donors, underscoring the prevalence of VL6-57 transcripts in human B cell repertoires. Analysis of published single-B V(D)J sequences also confirmed considerable expansion of B cells expressing VL6-57 antibodies with the paired “WLRG” motif after SARS-CoV-2 vaccination or infection. These findings suggest that naïve B cells expressing VL6-57 antibodies with the paired “WLRG” motif are activated after SARS-CoV-2 exposure, undergoing clonal expansion and class switching.
To confirm the reactivity of VL6-57 light chains from SARS-CoV-2 naïve donors with the SARS-CoV-2 RBD, 5 heavy chains containing the “WLRG” motif were paired with VL6-57 light chain to generate recombinant antibodies H4, H5, H14, H16, and H18. All 5 antibodies bound to the SARS-CoV-2 WT RBD, with H18 exhibiting cross-reactivity with RBDs of sarbecoviruses. Additionally, H18 showed neutralization activity against SARS-CoV-2 variants prior to Omicron BA.1 and had the ability to trigger S-protein conformational change and inhibit S-protein-ACE2 mediated cell-cell fusion. Structural analysis revealed that H18 binds to the convergent epitope targeted by VL6-57 antibodies through hydrophobic interactions in the HCDR3 and stabilizing hydrogen bonds in the LCDR1. These findings confirm that the VL6-57 light chain can effectively pair with multiple heavy chains to target the convergent epitope on SARS-CoV-2 spike RBD.
As observed, R1-26 and H18 lose their binding and neutralization activities against SARS-CoV-2 Omicron BA.1 variant. Structural analysis of VL6-57 antibodies revealed that residues S371, S373, and S375 are located in the conserved epitope, suggesting that the reduced activity of VL6-57 antibodies against Omicron BA.1 may be associated with the S371L-S373P-S375F mutations. Mutagenesis analysis demonstrated that revert mutations at residues 371, 373, and 375 on the Omicron BA.1 RBD completely restored the binding activities of R1-26 and H18. Additionally, structural comparisons of WT and Omicron BA.1 or BA.2 RBDs indicated that the backbone conformations at residues 373 to 375 are altered by the introduction of S373P and S375F mutations, implying that these changes in epitope residues likely impair binding by the VL6-57 antibodies.
This study identifies a class of VL6-57 light chain utilizing antibodies with the ability to pare with diverse heavy chains to target a convergent epitope defined by RBD residues S371-S373-S375. These antibodies are present in SARS-CoV-2-naïve individuals and clonally expand in COVID-19 patients. Omicron-specific mutations at S371, S373, and S375 enable immune escape from this antibody class, suggesting that immune pressure drives the emergence of these mutations, contributing to the antigenic evolution of SARS-CoV-2.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in