Antimicrobial resistance - a tale of silence and lessons
Published in Microbiology

The emergence and spread of antimicrobial resistance (AMR), especially through bacteria, is a global threat to public health due to the inability to comprehensively treat infections. Through horizontal gene transfer, bacteria can acquire antimicrobial resistance genes (ARGs) via conjugation using plasmids or through transduction employing phages. Therefore, AMR has been classified as the ‘silent pandemic’1 whereby it is no longer constrained by geographical or human-animal borders. Interestingly enough, Alexander Fleming warned of the misuse of antibiotics, which may lead to selection of resistant bacteria2. In order to understand the dissemination of antibiotic resistance, it is therefore necessary to characterize the resistome (the collection of ARGs in a given environment or organism) within different microbial reservoirs, and to unravel the extent to which the transfer of genes to and from de facto and putative human pathogens occurs. Importantly, the evolutionary timescales of AMR are critical to understand given the propensity for constant exchange of genetic material(s) within microbiomes which may drive rapid expansion of resistant taxa. In general terms, there exists an urgent need to understand the exchange of ARGs within and between biomes in the context of One Health and microbiome-borne pathogenic potential as it relates to human infectious and chronic diseases (Figure 1).
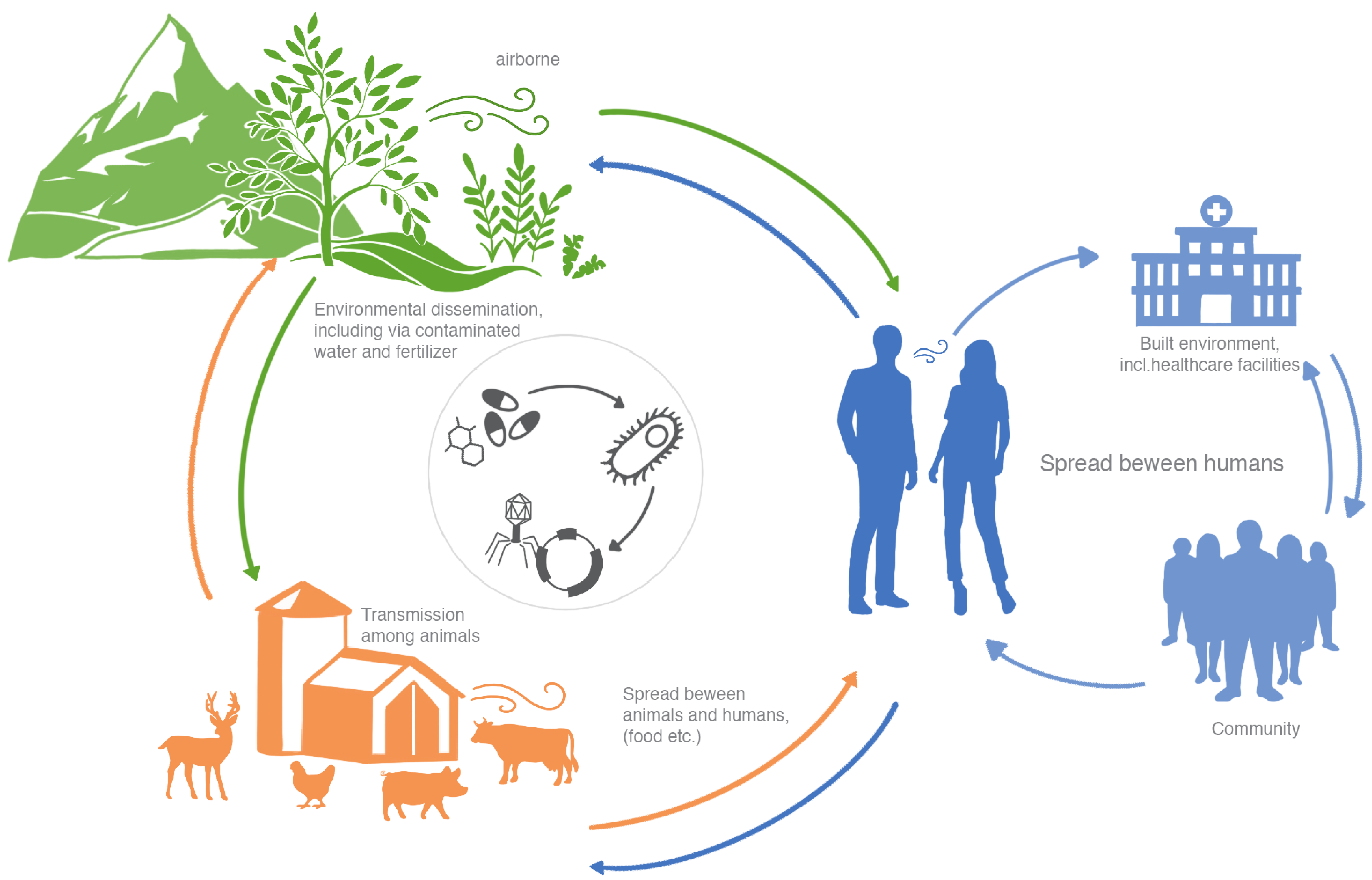
Figure 1: AMR dissemination in One Health. MGE-mediated (i.e. phage, plasmids and integrons) dissemination of AMR across different biomes.
While all of the above is meant to establish a scientific rationale for why we looked into the evolution of the murine gut resistome - the reality of our crusade, like most exciting studies in Science is - that it is not entirely true. Though the outcomes of our analyses indeed reveal the often overlooked role of integrons in ARG dissemination, the origins are more serendipitous culminating in a laundry list of lessons learnt about collaboration(s) and Science in general.
On a cold winter’s day in March 2019, the Microbes in One Health doctoral training unit at the University of Luxembourg was hosting a lecture about the challenges of controlling multidrug-resistant microorganisms at the European level. As fate would have it, we attended the lecture (mostly for the food after the fact). By the end of the seminar though, a question lingered as to whether the effect of antibiotic treatments on the microbiota in common practices in the animal and human microbiomes had been assessed. For example, pretreatment with antibiotics prior to fecal microbiota transplantation is a common process when treating colitis3. Similarly, antibiotic treatment is a proposed approach for both studying4 and modifying bacteria in the gastrointestinal tract5.
What ensued next is (maybe) lesson #1 for all graduate students and postdocs - perseverance! We first pitched the idea and a plausible hypothesis (you’ll have to read the article - Evolution of the murine gut resistome following broad-spectrum antibiotic treatment for this) to Paul (our co-PI). Despite having a lot of irons in the fire and potential for things to get overwhelming, there was an encouraging note from Paul to at least peruse the literature and determine if antibiotic treatment in microbiome-related studies was reported to assess AMR evolution in vivo. Lesson #2 - find yourselves (if possible) a supportive mentor who is also curious (what was to us) about “interesting questions”. Since we did not have an animal colony Paul pointed us to Elisabeth (co-PI) whose group studies colon cancer using murine models. And this, dear friends, is lesson #3 - collaborators and collaborations. This was our opening, and we pitched the idea to Elisabeth and her graduate student, Mina, who were equally intrigued that no studies till that date had reported AMR acquisition and/or evolution within the intestinal microbiome as a consequence of antibiotic treatment. They enthusiastically agreed to amend their animal protocols and sample the mice literally every day prior to and after treatment with the antibiotic cocktail (Figure 2). Importantly, our collective insights including their input allowed us to establish a clear experimental plan which would likely yield results.
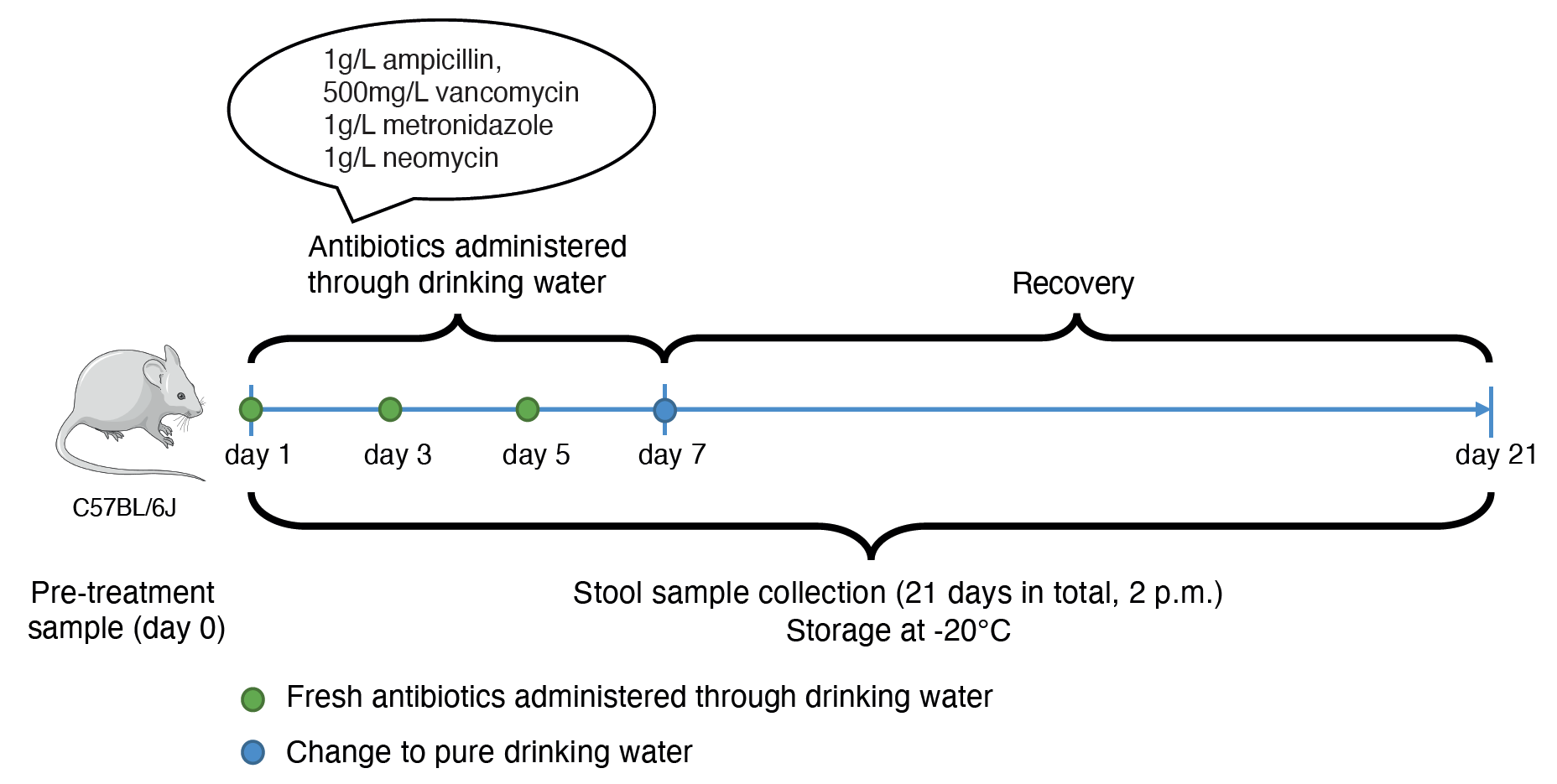
Figure 2: Longitudinal experimental design. Longitudinal fecal samples were collected from antibiotic-treated and untreated mice to assess the evolution of the resistome.
Fast-forward to eight months later and thanks to another ongoing pandemic (SARS-CoV-2), there was a shortage in sequencing kits delaying the study severely. Yet again lessons #1 and #3 played a major role along with the Sequencing Platform manager (Rashi) in what followed, and we eventually found that taxa from Akkermansiaceae and Enterobacteriaceae families acquired ARGs due to the treatment6. We then hedged all bets on phages being the primary carriers of ARGs since most plasmids would’ve been depleted along with the taxa as a consequence of the treatment. Alas, it wasn’t meant to be - we didn’t find a big difference between plasmids and phages, and something was amiss. Back to the drawing board it was and then it struck us that aside from plasmids and phages, MGEs are also constituted of integrons and transposable elements. While the pursuit of the latter was unfruitful, we found that integrons, without much fanfare, played a key role in ARG dissemination. In fact, we observed that integrons were instrumental in ARG acquisition both within Akkermansiaceae and Enterobacteriaceae (Figure 3). This was lesson #4 - the literature never disappoints in terms of ideas/hypotheses to test. The rest, as they say, is history!
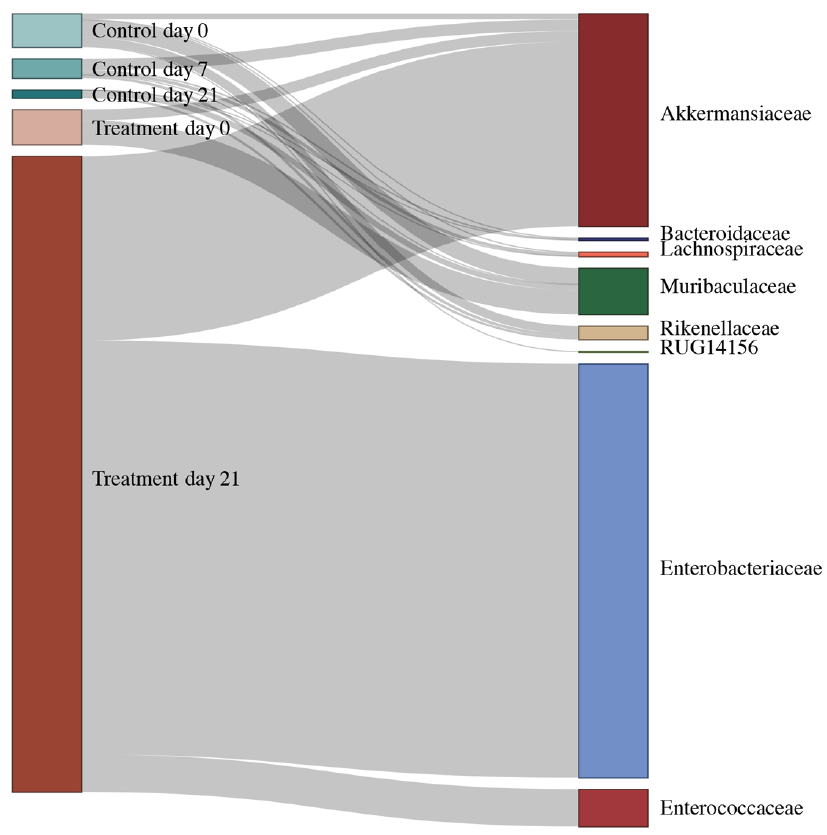
Figure 3: Integrons, the often-overlooked entities of AMR dissemination within the resistome. Alluvial diagram demonstrating the abundance of ‘complete’ integrons linked with observed antimicrobial resistance genes in specific taxonomic families.
Or is it?! The way we see it, this is only the beginning driven by lesson #5 - serendipity. Although rare and never manufactured, ‘Eureka moments’ do exist in Science. Our findings are a prime case-in-point to this end. Member genera from the Akkermansiaceae (eg. Akkermansia muciniphila) family, found in antibiotic-treated mice, have been key players, reported elsewhere, in improving human health7, and have also been implicated in chronic diseases including Parkinson’s8,9. We, therefore, now believe that it is time for due diligence and improved antibiotic stewardship, in murine models and medical treatment alike, given the longer-term disease-causing propensity of resistant bacteria such as Akkermansiaceae.
Co-authored by: Laura de Nies and Susheel Bhanu Busi (with input from all co-authors)
Image credits and copyrights: Laura de Nies
References
- Mendelson, M., Sharland, M. & Mpundu, M. Antibiotic resistance: calling time on the ‘silent pandemic’. JAC Antimicrob Resist 4, dlac016 (2022).
- Rosenblatt-Farrell, N. The landscape of antibiotic resistance. Environ. Health Perspect. 117, A244–50 (2009).
- Keshteli, A. H., Millan, B. & Madsen, K. L. Pretreatment with antibiotics may enhance the efficacy of fecal microbiota transplantation in ulcerative colitis: a meta-analysis. Mucosal immunology vol. 10 565–566 (2017).
- Lundberg, R., Toft, M. F., August, B., Hansen, A. K. & Hansen, C. H. F. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes 7, 68–74 (2016).
- Kennedy, E. A., King, K. Y. & Baldridge, M. T. Mouse Microbiota Models: Comparing Germ-Free Mice and Antibiotics Treatment as Tools for Modifying Gut Bacteria. Front. Physiol. 9, 1534 (2018).
- de Nies, L., Busi, S.B., Tsenkova, M. et al. Evolution of the murine gut resistome following broad-spectrum antibiotic treatment. Nat Commun 13, 2296 (2022).
- Dao, M. C. et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65, 426–436 (2016).
- Heintz-Buschart, A. et al. The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 33, 88–98 (2018).
- Bedarf, J. R. et al. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 9, 39 (2017).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in