As obligate intracellular parasites, viruses require host cell systems for the production of progeny viruses. This necessitates intricate interactions between viruses and host cells that often lead to significant intracellular reorganization. The overall goal of our research is to determine how viruses integrate with and manipulate host cell systems, and how mechanistic insights can lead to better understanding of both disease states and fundamental cellular processes. This research focuses primarily on flaviviruses, including Dengue virus and Zika virus, which, upon infection, alter the endoplasmic reticulum (ER) structure to form specialized membrane compartments serving as viral replication sites. Analysis of host proteins involved in forming or maintaining ER membrane structure revealed that Atlastin (ATL) GTPase proteins support flavivirus infection. Atlastins are membrane fusing factors that are involved in forming the three-way junctions in-between ER tubules, which are required for the production of the ER tubule network. This ER tubule network facilitates membrane connections throughout the cytosol and is required for many cellular processes. In our study, we found that two of the 3 mammalian ATL paralogues, ATL2 and ATL3, facilitate flavivirus infection. Interestingly, though these Atlastin paralogues are generally thought to have redundant functions, they are involved in different steps of the virus infection cycle; with the ATL2 paralogue supporting viral RNA replication and the ATL3 paralogue being required for the production of infectious virus progeny.
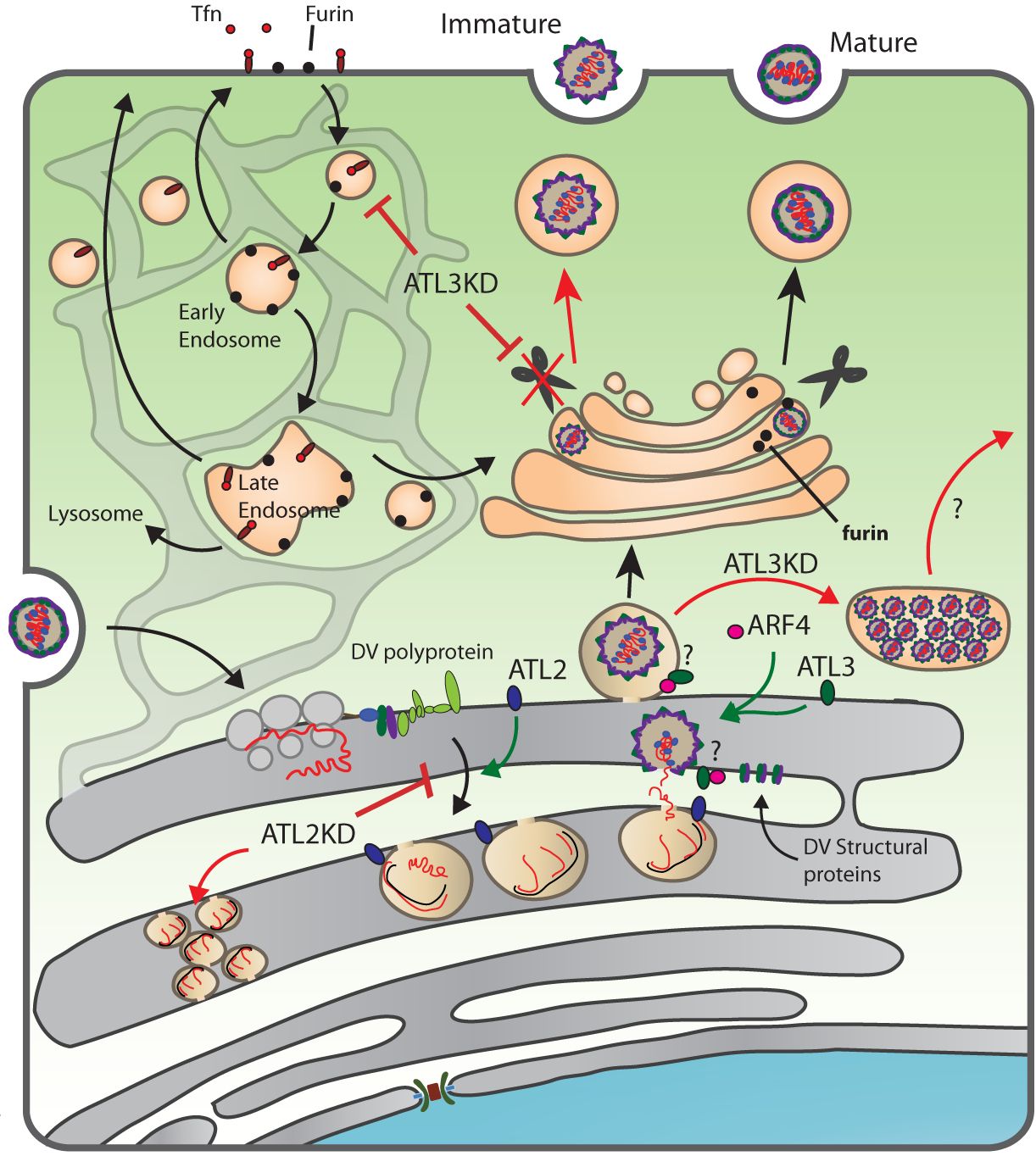
These two different effects on virus infection gave us a unique opportunity to not only study how these host factors are used by flaviviruses, but also to functionally characterize these different Atlastin paralogues. We approached this project from both the virus and cellular perspective with the goal of gaining broader insights into the functions of atlastins. Focusing on ATL3, we demonstrate a specific role in the maturation of flavivirus particles, a process that requires the cleavage of a viral envelope protein by the cellular protease furin, and obtained evidence linking ATL3 to endosomal recycling pathways.
Additionally, by taking advantage of the different viral phenotypes induced by selective ATL depletion, we used the virus as a tool to characterize ATL2 or ATL3 leading to the identification of domains that functionally differentiate these two host factors. Specific mutations in ATL1 or ATL3, but not ATL2, are linked to hereditary spastic paraplegia, a neurological disorder leading to muscle weakness and difficulty in walking. Our results as well as future analysis using these tools can give useful insights into the roles of atlastins in these diseases. From this work we are able to make conclusions on how ATLs function both in the context of flavivirus infection and in naïve hosts cells and demonstrate the value of using viruses as probes to unravel fundamental principles of cellular homeostasis.
This study was only possible through a productive collaborative effort between researchers and institutions. Thanks to Mirko Cortese, Pietro Scaturro, Berati Cerikan, Jeremy Wideman, Keisuke Tabata, Thaís Moraes, Olga Oleksiuk and Andreas Pichlmair for your contributions.
Written by Christopher Neufeldt and Ralf Bartenschlager
Link to paper: https://www.nature.com/articles/s41564-019-0586-3
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in