Atomic-resolution imaging of complex chiral molecular structures by machine learning based laser-induced electron diffraction
Published in Chemistry
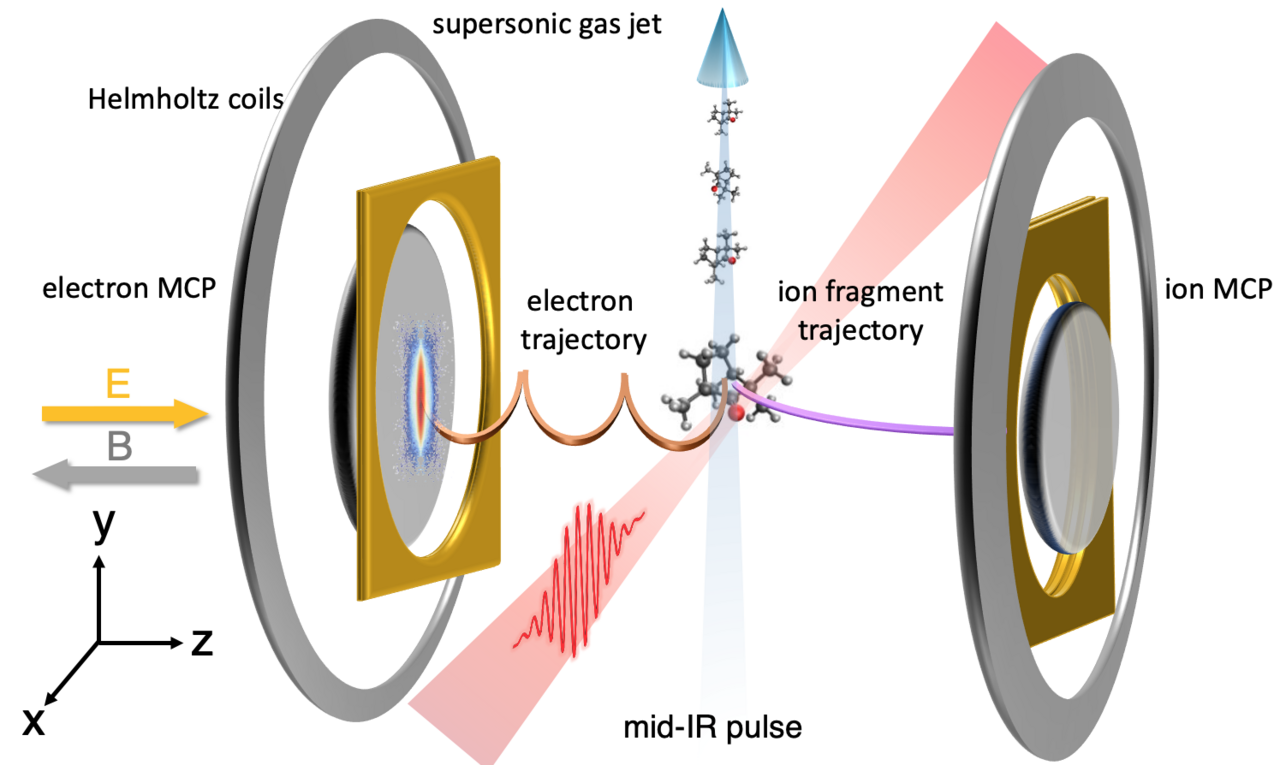
Until very recently, the idea of directly observing how molecules break, or transform, during chemical reactions was unfathomable. To provide revolutionary insights into the precise nature of the structural rearrangement such as transition states, proton migration, isomerization and nuclear dynamics in conical intersections, an imaging method that could achieve the atomic resolution is highly required. Laser-induced electron diffraction (LIED) [1,2] is a technique that allows to pinpoint the individual atoms inside a single molecule and capture the dynamics where the atoms move during a reaction. It is based on self-imaging the structure by recolliding a laser-driven attosecond wavepacket after photoionization with combined picometer and attosecond spatiotemporal resolutions. In 2016, our group evolved the LIED technique to image and track the molecular bond breakup in acetylene (C2H2), nine femtoseconds triggered by its ionization, a method coined “molecular selfie” [3].
Applying the LIED technique to take snapshots of small gas-phase molecules proved to be an extremely powerful tool to understand the intertwining of molecules and how they react, change, break, bend, etc. However, this technique was never applied to more complex molecular structures. A general challenge for diffraction-based imaging methods is the necessity to extract information from the measured diffraction patterns, which relies on locating a global extremum in a multi-dimensional solution space. The larger the molecule is, the harder the structural retrieval becomes. Also, it is necessary to calculate many thousands of molecular configurations for all possible orientations of the molecule, something that would take ages.
Machine learning (ML) is well-qualified to address such difficulties due to the simultaneous consideration of manifold degrees of freedom. Based on the ML-LIED framework, we prove the accurate retrieval of three-dimensional (3D) structures of large complex molecules.
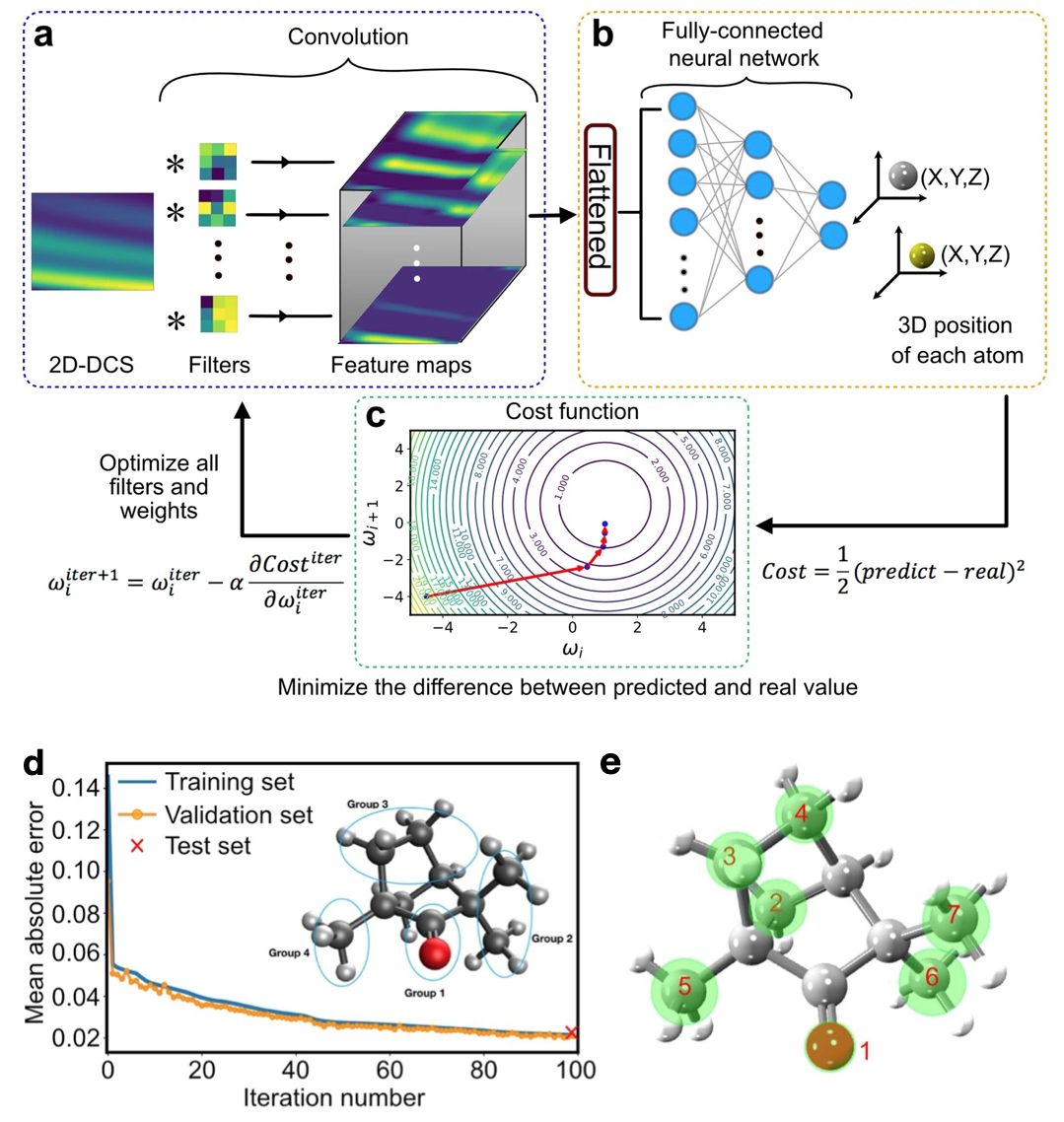
Fig. 1 Extracted (+)-Fenchone molecular structure. (a)The diffraction pattern (two-dimensional differential cross-section, 2D-DCS) is convolved with different filters to produce an ensemble of feature maps. (b) The feature maps are first flattened into a one-dimensional array and multiplied by the weights between each neuron of all layers in the fully-connected neural network to predict each atomic position in the molecule. (c) A schematic contour plot of the cost function with two weights. (d) Comparison between training and validation set for a subset of molecular voluminal. (e) Extracted (+)-Fenchone molecular structure of the measurement. Green circles represent uncertain areas.
After the preliminary examination of smaller 1D and 2D molecules (C2H2 and CS2), we test our ML framework to extract the structure of the chiral molecule (+)-Fenchone (C10H16O; 27 atoms) measured with LIED. For such large and complex 3D molecules, ML has the decisive advantage to interpolate and learn between coarse grids of pre-computed structures, taking into account manifold degrees of freedom in the solution space. Thus, we can establish a sufficiently reduced database that only considers (i) the four groups of atoms of the molecule (see the inset of Fig. 1d) and (ii) the global structural changes at the molecular scale. Next, we train our ML model with such a reduced database to find relationships between molecular structures and corresponding diffraction patterns (two-dimensional differential cross-section, 2D-DCS). This approach dramatically reduces computation time. The mean absolute error (MAE) (prediction error), defined by the absolute difference between the predicted and actual value, evaluates the model’s accuracy during the training process. Figure 1d shows the MAE achieved using the neural network at each iteration, convolved with the training and validation sets of simulated data. This achieves an MAE of 0.02 at the end. Also, a strong correlation between the experimental and predicted theoretical 2D-DCS is achieved with a Pearson correlation coefficient of 0.94. Figure 1e shows the 3D arrangement of the seven atoms of (+)-Fenchone (green circles). A slight deviation from the ML-LIED-measured structure and the equilibrium ground-state neutral molecular structure (red triangle) is involuntarily caused by the presence of the LIED laser field.
This result is of major importance because being able to calculate the 3D molecular structure of complex molecules with sufficient structural resolution has been, so far, a very difficult challenge to overcome. This study is a major step forward in this field, where the combination of LIED, machine learning and the CNN network, has not only shown the ability to predict and determine the structure of these large molecules, but also do it within a completely reasonable computing processing time. LIED combined with ML provides a new general solution to overcome standing problems and a new opportunity to determine the structure of large and complex molecules.
For more information, you can read more about our work in Communications Chemistry by following the link: https://www.nature.com/articles/s42004-021-00594-z
References
[1] T. Zuo, A.D. Bandrauk, P.B. Corkum “Laser-induced electron diffraction: a new tool for probing ultrafast molecular dynamics” Chem. Phys. Lett. 259, 313 (1996).
[2] C. Blaga et al. “Imaging ultrafast molecular dynamics with laser-induced electron diffraction”, Nature 483, 194–197 (2012).
[3] B. Wolter et al. “Ultrafast electron diffraction imaging of bond breaking in di-ionized acetylene,” Science 354, 308–312 (2016).
[4] X. Liu et al. “Machine learning for laser-induced electron diffraction imaging of molecular structures”, Comm. Chem. 4, 154 (2021).
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
f-block chemistry
Publishing Model: Open Access
Deadline: Feb 28, 2026
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in