
Although we sometimes want to look the other way on this topic, like it or not, we all derived from our parents, who at some point had sex, and they are a product of their parents, who also reproduced, and so on. It is mind-blowing to think that if just one of our ancestors (including bacteria, first eukaryotes and then mammals) failed to reproduce, we wouldn’t exist. Even more astounding is the realization that this continuous process started billions of years ago. We are all living fossils; basically, each and every one of us is billions of years old, with not a single moment in time where our DNA was not a part of a living being. We have evolved in the process not only in a physical way, but behavioral as well. Or so we like to think.
Apart from those who watched Game of thrones, have you ever asked yourselves, how do we choose mates and why do we avoid mating with our relatives? How disappointing, you watch a TV program for 8 years only to find out that your favorite two characters are in love AND quite closely related (aunt and nephew). How disappointing, right? We all know inbreeding can be dangerous for our genes, as it can result in a reduction of fitness of an inbreeding population and can lead to the harmful expression of deleterious recessive alleles (inbreeding depression)1,2. Okay, so inbreeding is bad. What about outbreeding and what is the optimal relatedness to produce offspring with the highest fitness? The results of a study from Iceland, drawn from all known couples of the Icelandic population born between 1800 and 1965, showed that the greatest reproductive success was observed for couples related at the level of third and fourth cousins and declined if couples were less (more distant cousins) or more related (first and second cousins)3. Similarly, animals are known to avoid inbreeding as well as outbreeding - they prefer mates of intermediate relatedness thereby avoiding genetic and somatic cost of outbreeding4.
So how is it possible, that even animals avoid –in and –out breeding, without understanding the dangers of both? How do they do it? It turns out that evolution took care of that by developing certain mechanisms to prevent inbreeding among related individuals, and that kin avoidance stems from kin discrimination, which is an ability to discriminate kin from non-kin and is important in mate choice as well as nepotism. If we over-simplify it, this can be done by learning the characteristics of siblings during immaturity and by phenotype matching, where an individual compares another individual’s cues to its’ own. The mechanisms of kin recognition are far more complex to be explained in detail here and can also depend on past social experiences of the animal (or human for that matter!).
It all sounds very complicated. So complicated, that one would think you have to be at least an animal (if not human) to be able to process all this information in order to identify kin from non-kin. But what if I told you, that we found evidence, that bacteria not only identify kin from non-kin, but also preferentially mate with a non-kin strain? Now when I say mate, I mean up-regulate genes for natural transformation upon meeting a non-kin strain, attack the non-kin strain and integrate it’s DNA. It’s not a very romantic exchange of DNA, but then again, think of spiders – where females eat males after copulation.
There has been controversy to why bacteria evolved natural transformation, which is, by the way, the only horizontal gene transfer mechanism entirely encoded on the bacterial chromosome. This suggests that it probably evolved to benefit the bacterium. Is it possible that it serves as a prokaryotic version of sex? This topic remains extremely controversial as it is still uncertain whether transformation evolved to maximize diversity and adaptation, to preserve/repair the existing genes5,6, or it serves as a means to uptake and use DNA for food 7. All three theories can be somewhat backed-up by scientific evidence.
Bacillus subtilis, a model Gram positive bacterium is naturally competent for transformation, meaning it readily takes up DNA and integrates it in it’s genome. There is strong evidence that transformation in Bacillus subtilis decreases dramatically with sequence divergence indicating that bacteria more frequently uptake DNA isolated from closely related microorganisms, this is also called sexual isolation8. This would benefit the bacterium if it needed DNA to repair it’s genes, but would bring very little new genetic information into the genetic pool (considering clonal growth of bacteria, no new info is obtained). However, experiments in the past addressing sexual isolation were most often performed by adding extracted DNA to a population of cells grown in an Erlenmeyer flask monoculture depriving the experimental design of intercellular interactions. In our recent research we discovered that bacteria can and will preferentially mate with a non-kin conspecific if allowed to interact with one another. Specifically, when they swarm on agar plates, they meet another swarm and form a boundary if they recognize it as a non-kin swarm (Figure 1). What follows is a mutual attack, but the outcome depends on which strain carries which attack and defence genes and usually one strain wins and the other – dies. In this process, the attacker up-regulates genes involved in competence for transformation so that it can uptake and integrate DNA more efficiently. Surprisingly, the presence/absence of attack and defense genes correlates to genetic relatedness – this means the more they are related, less likely it is they will harm each other. Makes sense, right?
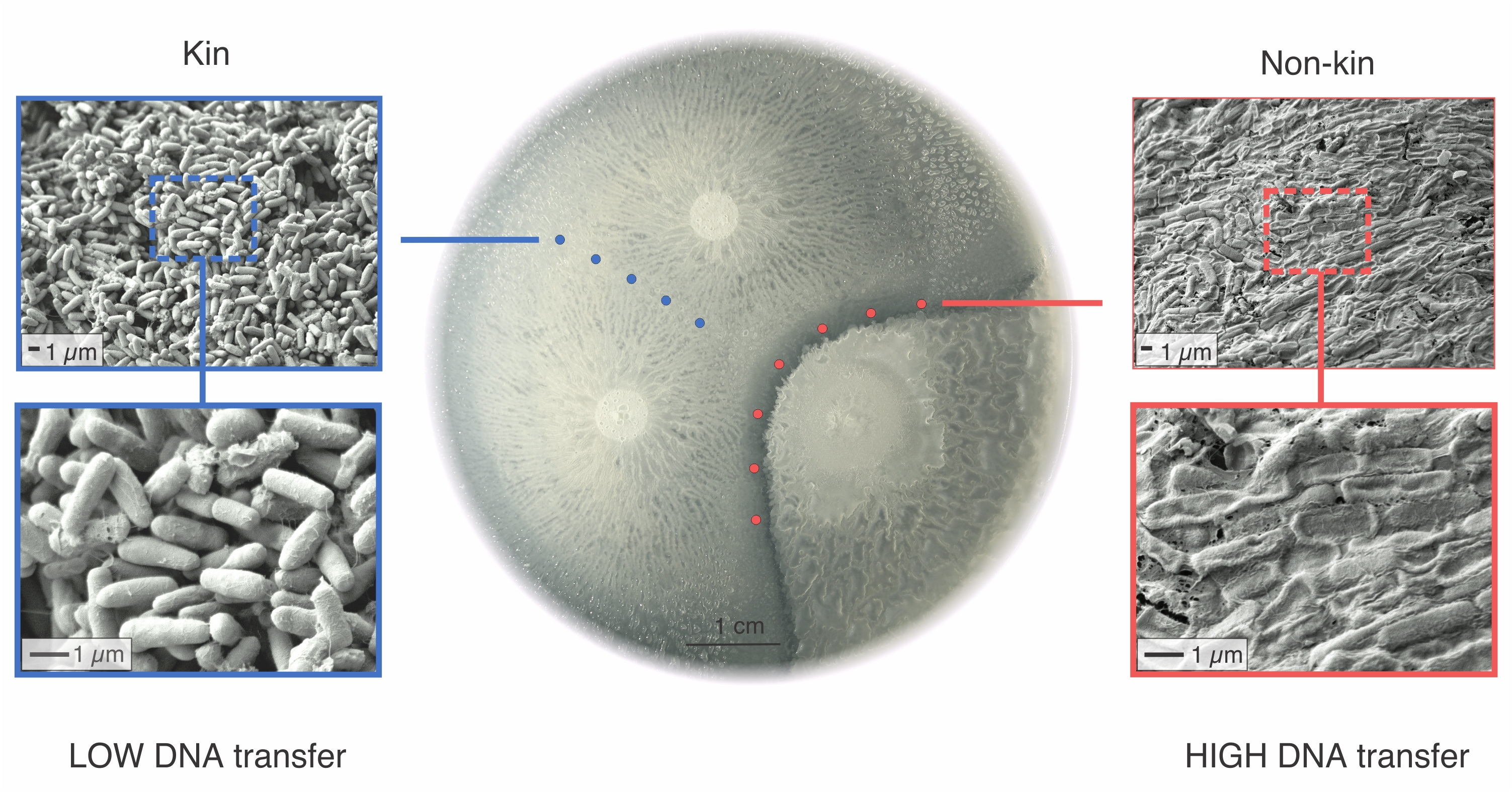
It is most likely, that bacteria developed kin discrimination to protect it’s kin and attack less related non-kin strains. But it is astounding that evolution coupled kin discrimination with genetic exchange, promoting intermediately related DNA transfer, which can bring novel genes, but is still safe as it comes from the same species or even niche. Sexual isolation on the other hand makes sure that divergent DNA originating from distinct species is not integrated. And this is actually the concept of species that is so easy to understand when we think of animals: species are groups of actually or potentially interbreeding populations which are reproductively isolated from other such groups9.
If we return humans, we also discriminate before we mate, and sometimes we are not very smart in our choices. Nevertheless, humans don’t just mate for reproduction but for fun also and one thing is for sure: those who fight, exchange DNA more efficiently, be it humans or bacteria.
For the full details, check our paper at https://www.nature.com/articles/s41467-021-23685-w
1 Charlesworth, B. & Charlesworth, D. The genetic basis of inbreeding depression. Genetical research 74, 329-340, doi:10.1017/s0016672399004152 (1999).
2 Pusey, A. E. in The International Encyclopedia of Anthropology 1-6 (2018).
3 Helgason, A., Palsson, S., Gudbjartsson, D. F., Kristjansson, T. & Stefansson, K. An association between the kinship and fertility of human couples. Science 319, 813-816, doi:10.1126/science.1150232 (2008).
4 Pusey, A. & Wolf, M. Inbreeding avoidance in animals. Trends in Ecology & Evolution 11, 201-206, doi:https://doi.org/10.1016/0169-5347(96)10028-8 (1996).
5 Michod, R. E., Wojciechowski, M. F. & Hoelzer, M. A. DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics 118, 31-39 (1988).
6 Claverys, J. P., Prudhomme, M. & Martin, B. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu Rev Microbiol 60, 451-475, doi:10.1146/annurev.micro.60.080805.142139 (2006).
7 Redfield, R. J. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered 84, 400-404 (1993).
8 Majewski, J. Sexual isolation in bacteria. FEMS Microbiol Lett 199, 161-169 (2001).
9 Mayr, E. Systematics and the Origin of Species. (New York: Columbia University Press, 1942).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
I really enjoyed this blog post...it made me laugh. And very cool paper too!