Bacterial oxylipins: a key to multicellularity and to combating antimicrobial resistance?
Published in Microbiology

Oxylipins serve as cell-to-cell communication signals to many multicellular eukaryotes. In vertebrates, 20-carbon oxylipins called eicosanoids orchestrate inflammation, an ancient and stereotypic response of our tissues to any injury or pathogen invasion. Plants have no inflammation, but their oxylipins coordinate stress and immune reactions similarly to the way our eicosanoids do. Fungi use oxylipins for quorum sensing and regulation of reproduction, and brown algae use chemically peculiar oxylipins as pheromones.
In all of the aforementioned organisms, oxylipins are synthesised by evolutionary related enzymes. For instance, all of them use lipoxygenases — conserved enzymes catalysing PUFA peroxidation. In humans, this peroxidation leads to formation of leukotrienes and lipoxins. In plants, it leads to formation of jasmonates. In fungi, it leads to formation of quorum sensing oxylipins. The only described function of lipoxygenases is to form oxylipins. So, if we know that some organism has a lipoxygenase, we can say it can synthesise oxylipins. But for some organisms, we don't know what they do it for.
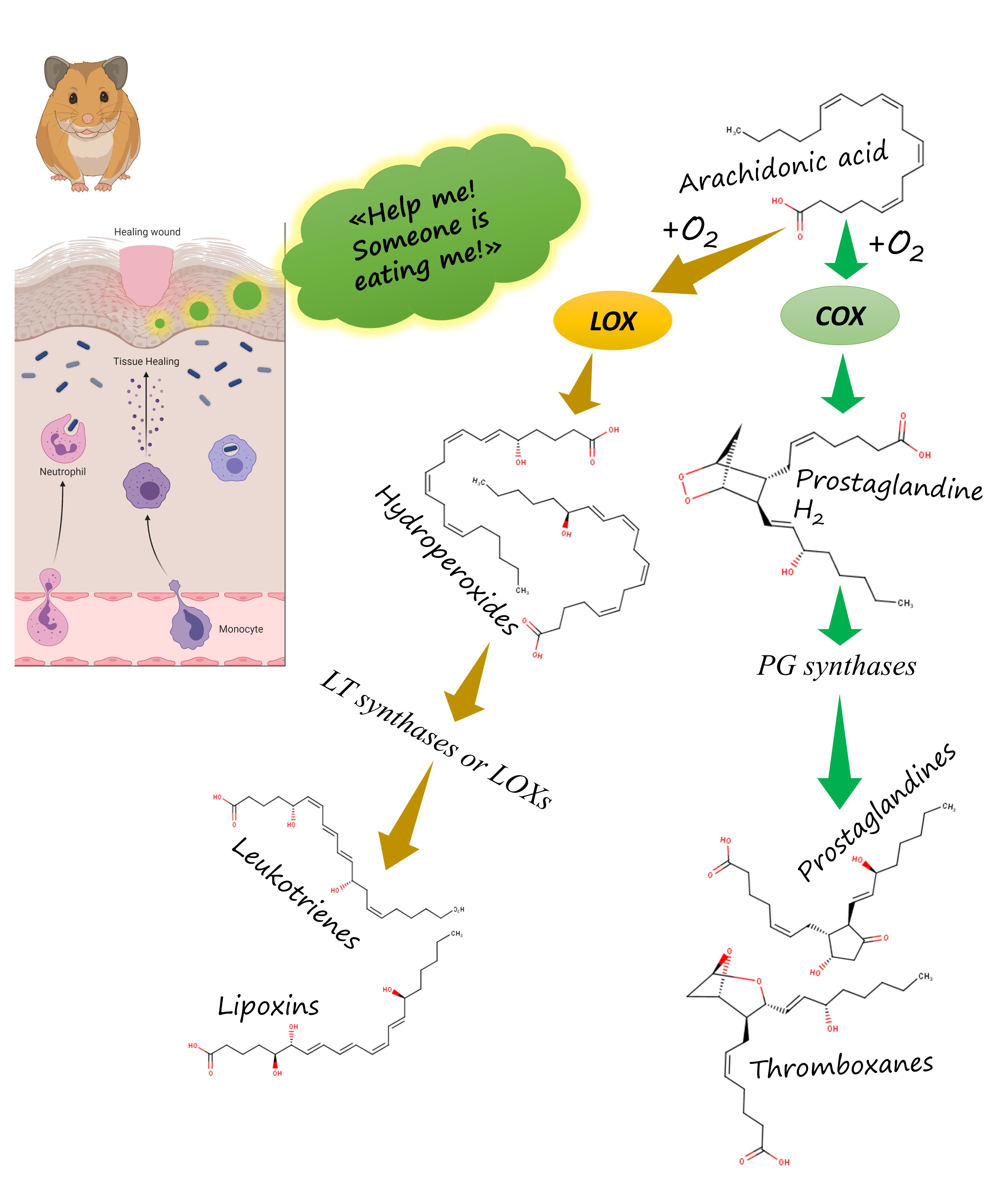
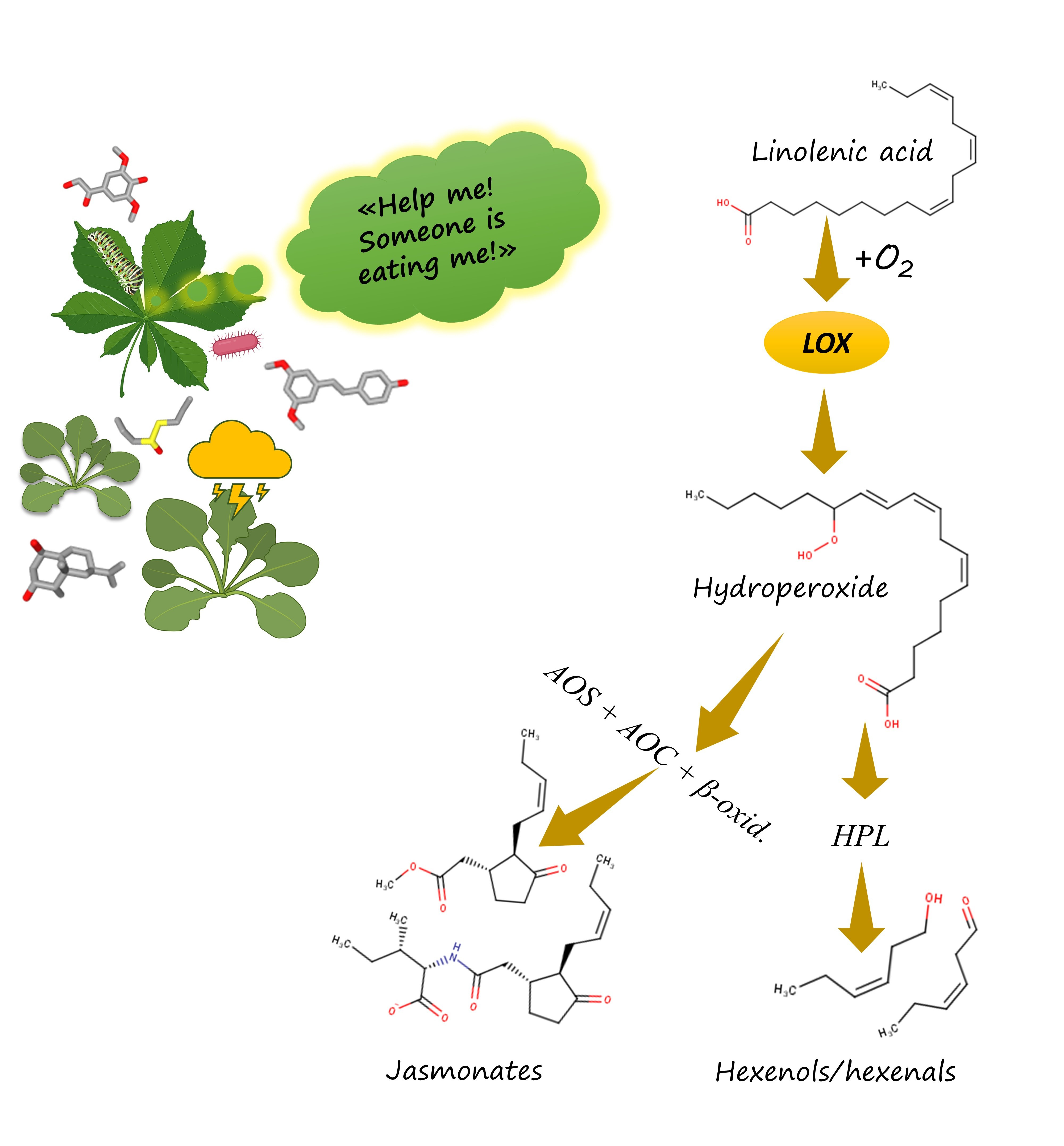
The further biosynthetic steps differ from human ones – for instance, hydroperoxide lyase (HPL) splits plant oxylipins to form volatile derivatives. Plants have no inflammation. Instead, they react to injury by releasing bactericidal, fungicidal and insecticidal compounds – phytoalexins. However, it is noteworthy that these quite dissimilar reactions on an injury on a pathogen invasion are triggered by lipoxygenase-derived oxylipins like our inflammation. It is a striking similarity across millions of years and different forms of multicellular beings. But what is the evolutionary origin of this similarity? This question is difficult to answer at once. Created with BioRender.com
Let's get together
It is the case of almost all bacteria. We know some of them – approximately 0.5% of all bacteria — have lipoxygenases. Experiments with some filamentous cyanobacteria and myxobacteria have shown they use lipoxygenase to form oxylipins, too, but the functions of these oxylipins remained unclear. It is interesting that both filamentous cyanobacteria and myxobacteria possess primitive multicellularity. Maybe bacterial cells communicate with oxylipins like animal or plant cells? Unfortunately, experimental data on this point are very scarce. We know only that:
1) cells of cyanobacterium Nostoc punctiforme 'cry' with oxylipins upon injury (like plant, animal, or algal cells do) [1]. But, in contrast to the cases of plants or animals, we don't know which response follows after this 'crying';
2) in contrast, oxylipins promote twitching motility in myxobacterium Myxococcus xanthus [2], but we don't know which stimulus triggers their release.
These two incomplete experimental evidences are very scarce regarding even the amount of known multicellular bacterial species. We tried to recompense this deficiency with a bioinformatic research. This way, we were able to process significantly larger volumes of data. We used lipoxygenases — easily recognizable by sequence — as markers of oxylipin biosynthetic ability. The results of our research have been published in Biochemistry (Moscow) [3].
We have found that lipoxygenases occur more frequently in 'multicellular' bacterial taxa – Cyanobacteria and Myxobacteria — than in unicellular taxa. And when we added a dataset of eukaryotic lipoxygenases to the bacterial ones and analyzed the lipoxygenase phylogeny, we have revealed the more convincing trend. From the beginning of time, lipoxygenases were transmitted rather by horizontal gene transfer than by vertical heredity. Saying simply, the lipoxygenase evolution is slightly similar to inheritance 'from a father to a son', rather it is similar to an 'epidemic' of gene transfers at the evolutionary timescale – a series of independent acquisitions by horizontal gene transfer events. The most important is our finding that independent lipoxygenase acquisitions coincided with emergence of multicellularity in different lineages. It means that lipoxygenases (and, thus, oxylipins) really show an evolutionary association with multicellularity.

Similarities between the evolution of lipoxygenases (as reconstructed by us) and the evolutions of other proteins, crucial for multicellularity, solidify this notion. Our reconstruction shows that oxylipin signalling evolution might start in cyanobacteria – maybe the first 'inventors' of multicellularity. In this respect, oxylipin signalling evolution is similar to the evolution of programmed cell death as discussed by Nick Lane in his well-known feature 'Origins of Death' for Nature [4]. We have discussed this similarity in detail in our popular synopsis 'All together now' published by the RSB's magazine The Biologist [5].
But later, when this article was being prepared for print, we found a new similarity. This autumn, a group of our colleagues has published an Invited Review 'Evolution of glutamatergic signaling and synapses' in Neuropharmacology [6]. In particular, authors showed coincidence between independent acquisitions of NTD domain in ionotropic glutamate receptors (iGluRs) and emergence of multicellularity in different eukaryotic lineages. It is very similar to lipoxygenase acquisitions coinciding with multicellularity emergence described by us. On of the authors of this article, Mikhail Nikitin, finds this similarity striking as well:
'When I saw your lipoxygenase phylogeny, it seemed to me that I see our own work on iGluRs,'
Mikhail said to us at the MCCMB'21 conference in Moscow this year.
These similarities raise an intriguing question: could oxylipins contribute to the origin of multicellularity at the Earth? This question remains open and very complicated since, even if oxylipins have contributed to 'bringing cells together', they are surely not crucial to be multicellular. Prof. Hartmut Kühn, Head of the Lipoxygenase Research Laboratory in Charité, commented our assumption:
'This hypothesis is interesting since multicellular assemblies of living cells require mechanisms of communication, and lipoxygenases might contribute. But obviously, they are not essential for intercellular communication. Even in invertebrates, which contribute more than 95% to all multicellular organisms on Earth, the percentage of lipoxygenase-containing species is lower than 1%. Clearly, multicellular life is possible without LOXs'
We hope to solve the complicated question in our further research. But it is not the only challenge of our research.
The Achilles' heel of antibiotic-resistant bacteria?
Primitive multicellularity was not the only common trait of lipoxygenase-carrying bacteria we identified. We were surprised to find the second subset of bacteria in our dataset — those are not multicellular, but share some common traits in terms of host-microbe relationships. Firstly, a lot of them are opportunistic pathogens affecting immunocompromised persons, hospital inpatients or people with specific diseases — for example, wtih cystic fibrosis. Secondly, a lot of them are versatile pathogens affecting both plants and vertebrates. More interestingly, some of them are plant symbionts or associated with specific marine invertebrates. And thirdly, a lot of these bacteria exhibit broad antibiotic resistance.
Now we are performing a more detailed analysis of this group. We hope it could help to understand the way these bacteria interact with a host's immune system and the secret of their versatility. One of these bacteria is already experimentally characterized – Pseudomonas aeruginosa uses oxylipins to evade human immune response [7]. Our research shows that it is not the only bacterium to make such 'oxylipin tricks': this mechanism could be exploited by a larger number of dangerous opportunistic pathogens. This fact increases the extent of the problem.
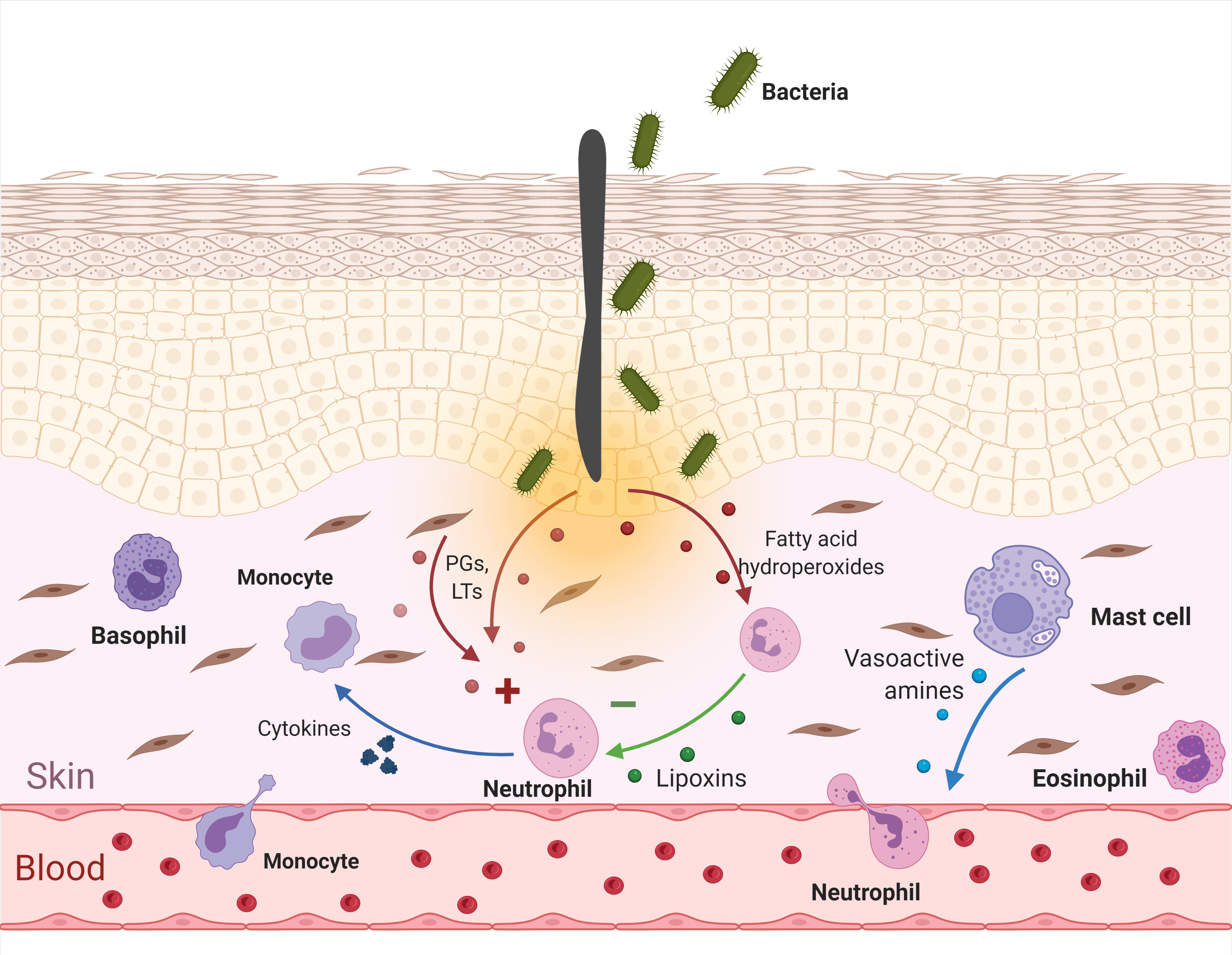
But oxylipins form not one-way signalling system. Rather, they form two regulatory loops counterbalancing each other. Injured cells release not only definite forms of pro-inflammatory PGs and LTs, but also fatty acid hydroperoxides and other compounds which can be further converted by lipoxygenases of leukocytes (being already present in the tissue) into anti-inflammatory oxylipins. There are different groups of them. Here, for clarity, I depict only lipoxins since we will need them in the further discussion. Prostaglandins and leukotrienes stimulate ('+') functions of neutrophils and monocytes (acting as 'help me' signals), and lipoxins suppress ('–') them (acting as 'attack over' signals). Due to this control, a normal inflammatory reaction is self-limiting. Created with BioRender.com
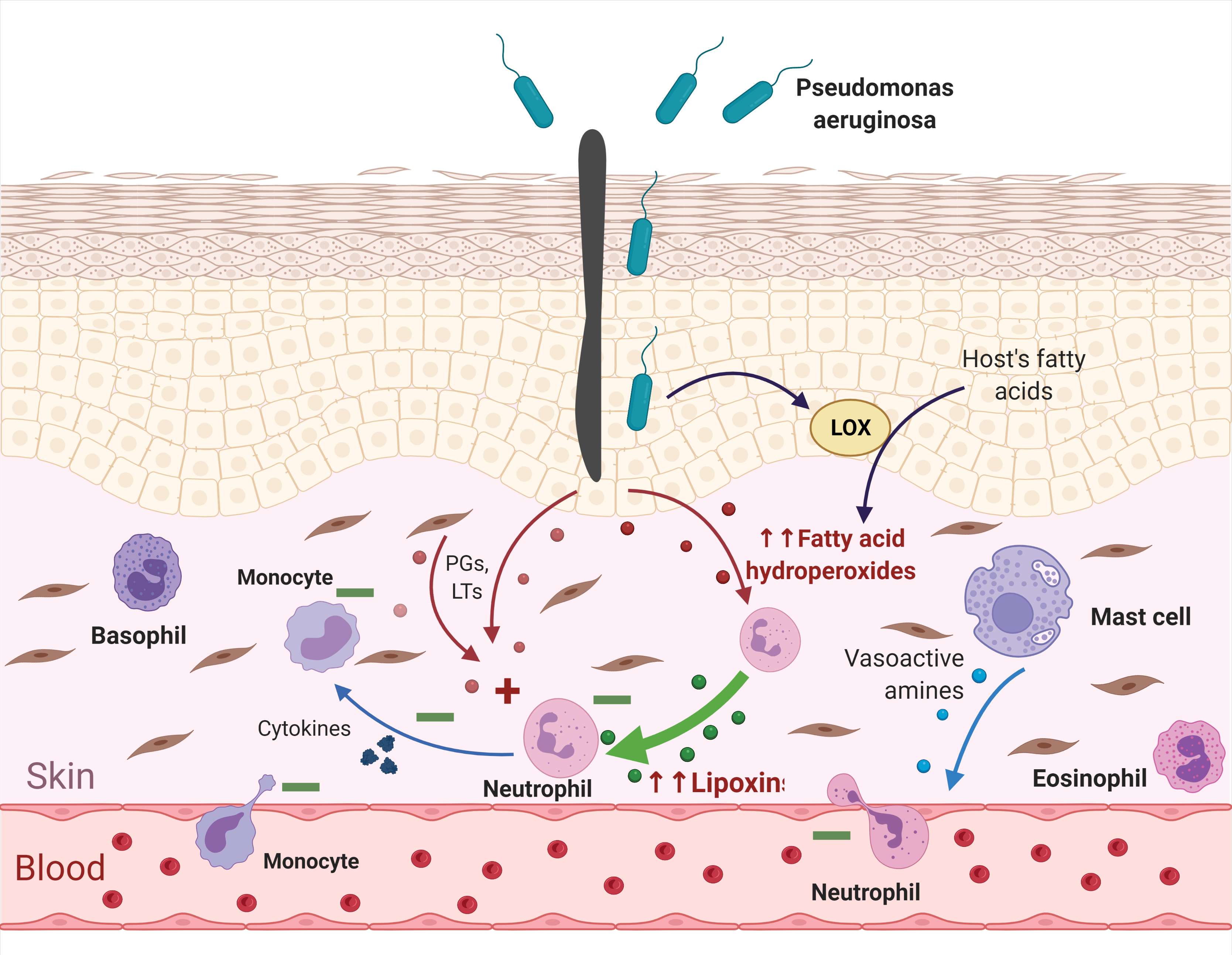
You can find a kind of 'pathogen blacklist' below — we suspect them in using lipoxygenase-derived oxylipins for immune response suppression. The species that have been added to our dataset since the time of publication in Biochemistry (Moscow) are given in bold italic. These updates are coming from updates of information in genomic and protein databases. So, it is an updated and shortened version of the table from our paper, where we have included only bacteria that are dangerous to humans.
| Order | Pathogens of concern |
|
Burkholderiales |
Burkholderia thailandensis, Burkholderia gladioli, Burkholderia singularis, Burkholderia stagnalis |
| Corynebacteriales | Mycobacteroides abscessus, Nocardia pseudobrasiliensis, Nocardia brasiliensis |
| Enterobacterales | Pluralibacter gergoviae, Cedecea lapagei, Pantoea ananas, Moellerella wisconsensis |
| Pseudomonadales | Pseudomonas aeruginosa |
The aforementioned facts raise a question: could lipoxygenases be the targets for developing new antibiotics? Despite relatively narrow action spectrum (which may be predicted by the table above), this approach may be winning by several reasons:
- many of these bacteria possess a broad antimicrobial resistance. So, their small number is more than counterbalanced by a danger they may pose;
- anti-virulence strategies are promising in developing new antibiotics, and blocking oxylipin signalling might be suited well to this conception;
- lipoxygenases are well studied in terms of binding site structure: blockers of human lipoxygenases are known, as well as crystal structures of Pseudomonas aeruginosa lipoxygenase. Our ongoing bioinformatic project could provide valuable insights into homology modelling of other pathogens' lipoxygenases.
These considerations have encouraged us to write a feature article in Laboratory News titled 'Microbial lipoxygenases: a next target against antibiotic resistance?' [8]. First of all, I would like to correct some assumptions from this article. Now we are happy to say that our alarm claims about 'superspreading' lipoxygenases between pathogens seem not to be confirmed now: genomic database update has deleted sequences closely related to Pseudomonas aeruginosa lipoxygenase from the databases. Now we may downgrade the alert, if I could express this way.
But it is not a reason for unawareness – the overall take-home message of the article is still actual:
'<...> We hope to attract the attention of our scientific and medical colleagues to this enzyme: maybe, it is the next target in fighting antibiotic resistance,'
we wrote in the conclusion of our article.
This conclusion has something in common with similar outreach articles of another scientific group which studies bacterial oxylipin signalling. The team led by Javier Campos-Gómez, Research Biologist in Southern Research, has recently discovered that oxylipins mediate cell-to-cell communication in Pseudomonas aeruginosa [9]. Earlier, they had proposed the model of intraspecific communication of Pseudomonas aeruginosa based on oxylipins [10]. Javier Campos-Gómez has already published the post in Nature Microbiology Community, where he summarises the findings of his team.
It is interesting that the oxylipin communication system described by Campos-Gómez's group is the first such system that is fully experimentally characterised. And, more interesting, it is based on oleate diol synthase (ODS) enzymes which are not homologous to our lipoxygenases and cyclooxygenases. It is the reason why it has appeared to be out of our view.
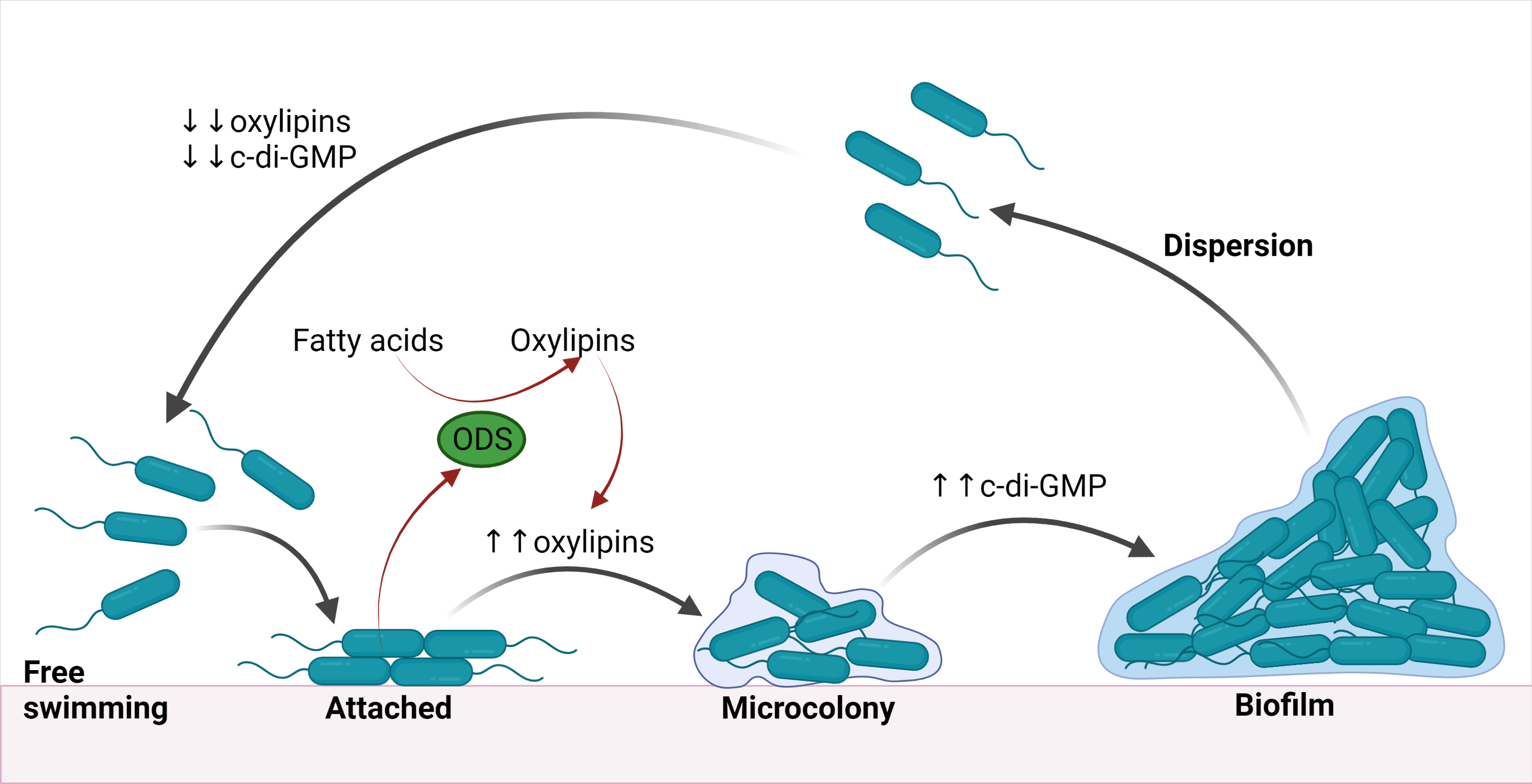
But the most interesting is the fact that the Campos-Gómez's group and we have arrived at very similar practical conclusions in such different ways. In an outreach article in Infection Control Today [11], Javier Campos-Gómez says:
'This is a step forward. Now, we know more about how oxylipins function in P. aeruginosa. They are signaling molecules involved in regulating biofilm formation and virulence. <...> This is important because we can design a new generation of antibiotics that target this oxylipin pathway'
And it is almost the same that we can say about our findings on the quite different oxylipin pathway and almost the same we tried to promote in our Laboratory News article. This surprising similarity seems to convince us that designing new antibiotics targeting bacterial oxylipin signalling is a really promising research direction.
What comes next?
The pathogenic action of bacterial lipoxygenases is now the main focus of our research. For us, it was an accidental finding to a great extent. But, given its great significance, we are now studying the evolutionary association between lipoxygenases, pathogenicity and symbiosis. Need to update genomic database information increases the significance of re-evaluating working hypotheses and danger grades for different pathogens. Therefore, we are now planning new publication(s) on the lipoxygenases of pathogenic bacteria — and, of course, new posts here based on new papers. Stay tuned!
And Happy New Year to everybody!
Acknowledgements
I acknowledge Anna Samoukina and Nadezhda Potapova as co-authors of our paper, without whose invaluable help it could not come out. And I say many thanks to biological artist Anastasiia Samoukina (Student, Program of Psychology, Neuroscience and Human Sciences, University of Pavia, Italy) for kindly providing us with the figures. Some other figures were created with BioRender.com.
All direct citations were used with a kind permissions of cited persons or taken from published sources.
Original paper
Read our paper on Springer Link (subscription access) or by this SharedIt link (free access).
Supplementary Materials
You could also get acquainted with a poster graphically outlining our paper. Besides this, I invite all colleagues to hear the record of my lecture 'Oxylipins: A Cry for Help' that was delivered online at the RSB London Branch on Feb 7, 2022.
Citation
This post was translated into Russian. This translation was formally published in Priroda journal with a reference to the original version in this blog and in compliance with the rules of Communities actual at the date published.
If you find this post interesting and would like to cite it in your article, you could cite this Russian version:
- Kurakin, G.F. (2022). Bacterial Oxylipins: a Key to Multicellularity and to Combating Antimicrobial Resistance? Priroda, (2), 26-32. DOI: 10.7868/S0032874X2202003X
The full information about the published version and its citation is available in English at the platform of Eco-Vector publishing house.
References
- Lang, I., & Feussner, I. (2007). Oxylipin formation in Nostoc punctiforme (PCC73102). Phytochemistry. Vol. 68, No. 8, pp. 1120–1127. https://doi.org/10.1016/j.phytochem.2007.02.028
- An, J.-U., & Oh, D.-K. (2018). Stabilization and improved activity of arachidonate 11S-lipoxygenase from proteobacterium Myxococcus xanthus. Journal of Lipid Research. Vol. 59, No. 11, pp. 2153–2163. https://doi.org/10.1194/jlr.m088823
- Kurakin, G. F., Samoukina, A. M., & Potapova, N. A. (2020). Bacterial and Protozoan Lipoxygenases Could be Involved in Cell-to-Cell Signaling and Immune Response Suppression. Biochemistry (Moscow). Vol. 85, No. 9, pp. 1048–1063. https://doi.org/10.1134/s0006297920090059
- Lane, N. (2008). Marine microbiology: Origins of Death. Nature. Vol. 453, No. 7195, pp. 583–585. https://doi.org/10.1038/453583a
- Kurakin, G. F., Samoukina, A. M., & Potapova, N. A. (2021). All together now. The Biologist, online version. https://thebiologist.rsb.org.uk/biologist-features/are-oxylipins-the-key-to-multicellularity
- Moroz, L. L., Nikitin, M. A., Poličar, P. G., Kohn, A. B., & Romanova, D. Y. (2021). Evolution of glutamatergic signaling and synapses. Neuropharmacology. Vol. 199, p. 108740. https://doi.org/10.1016/j.neuropharm.2021.108740
- Morello, E., Pérez-Berezo, T., Boisseau, C., Baranek, T., Guillon, A., Bréa, D., Lanotte, P., Carpena, X., Pietrancosta, N., Hervé, V., Ramphal, R., Cenac, N., & Si-Tahar, M. (2019). Pseudomonas aeruginosa Lipoxygenase LoxA Contributes to Lung Infection by Altering the Host Immune Lipid Signaling. Frontiers in Microbiology. Vol. 10. https://doi.org/10.3389/fmicb.2019.01826
- Kurakin, G. F., Samoukina, A. M., & Potapova, N. A. (2021). Microbial lipoxygenases: a next target against antibiotic resistance? Laboratory News. Online version. https://www.labnews.co.uk/article/2031213/microbial-lipoxygenases-a-next-target-against-antibiotic-resistance
- Martínez, E., Cosnahan, R. K., Wu, M., Gadila, Shiva. K., Quick, E. B., Mobley, J. A., & Campos-Gómez, J. (2019). Oxylipins mediate cell-to-cell communication in Pseudomonas aeruginosa. Communications Biology. Vol. 2, No. 1. https://doi.org/10.1038/s42003-019-0310-0
- Martínez, E., & Campos-Gómez, J. (2016). Oxylipins produced by Pseudomonas aeruginosa promote biofilm formation and virulence. Nature Communications. Vol. 7, No 1. https://doi.org/10.1038/ncomms13823
- New Bacterial Signaling Language Offers Pathway to Treat Infections. Infection Control Today. Online version. https://www.infectioncontroltoday.com/view/new-bacterial-signaling-language-offers-pathway-treat-infections
Post update log
- UPD from 08.05.2023: the lecture information updated.
- UPD from 10.05.2023: citation information added.
- UPD from 25.07.2024: citation information updated.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in