Beware, HER2-Low Breast Cancer. We See You.
Published in Cancer, Genetics & Genomics, and General & Internal Medicine
Not all breast cancers are the same; instead, there are various types, each with distinct characteristics. For years, immunohistochemistry (IHC) has been used to track breast cancer biomarkers, like HER2. On the heels of the DESTINY-Breast04 trial (NCT03734029), the antibody-drug conjugate (ADC) therapy trastuzumab deruxtecan (T-DXd) was approved for HER2-low breast cancer, highlighting the need to distinguish HER2-low from HER2-negative patients. In conventional HER2 classification, high HER2 means “positive,” treatable with T-DXd, whereas low or no HER2 means “negative.” However, ambiguous HER2-low tumors—scoring 1+ or 2+ on IHC with negative in situ hybridization—remained difficult to diagnose. Compounding this problem is the low reported concordance among pathologists in distinguishing between HER2 0 and 1+ IHC scoring for breast cancers. To overcome these hurdles, our team of pathologists, bioinformaticians, doctors and biologists at BostonGene set out to develop a transcriptomic-based solution for HER2-low detection.
The new kid on the block
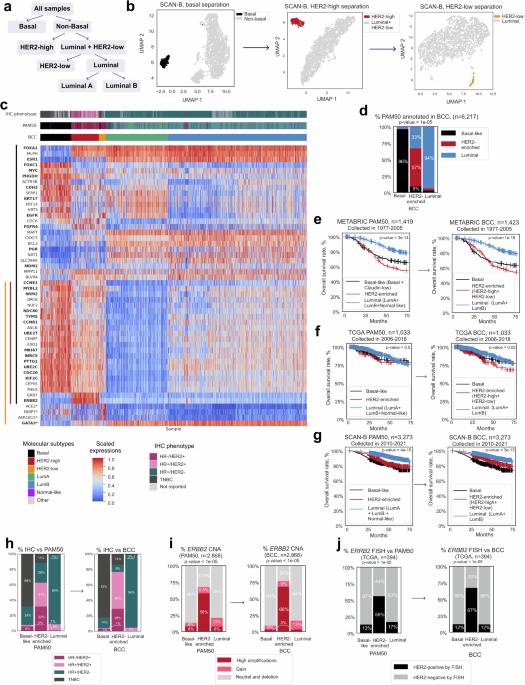
The 50-gene PAM50 test sorts breast tumors into Basal-Like, HER2-Enriched, Luminal A, Luminal B, and Normal-Like groups. Based on gene expression patterns in tumor samples, this approach yields a more precise classification than IHC does. However, emerging evidence from clinical trials suggests that PAM50-classified HER2-enriched tumors are not identical to those with ERBB2 amplification or high HER2 expression by IHC. The PAMELA (NCT01973660) and KEYRICHED-1 (NCT03988036) studies, for instance, showed the best treatment outcomes from HER2-targeted therapy on HER2-enriched tumors that presented with strong ERBB2 expression. Unfortunately, tumors designated as HER2-enriched by PAM50 can sometimes behave more like Luminal or Basal-Like tumors, complicating therapeutic decisions.
There’s another caveat: PAM50 wasn’t designed to detect HER-low tumors. The PAM50 classifier was published in 2015 and T-DXd gained approval for HER2-low breast cancers in 2022. So, what’s so special about these tumors that we now have new treatments for them?
A new tool for detecting HER2-low tumors
Our quest began with an attempt to answer some pressing questions. For instance, how do we overcome the inherent inconsistencies of IHC scoring? How do changes in ERRB2 expression impact the detection of HER2-low tumors? Leveraging our capabilities in next-generation sequencing and machine-learning algorithms, we determined that RNA sequencing (RNA-seq) analysis was the most practical approach. Hence, the Breast Cancer Classifier (BCC) was born.
The BCC is a platform-independent, transcriptome-based classification tool. It ranks the expression levels of a set of 63 genes in breast tumors in order to categorize them into five different categories—Basal, HER2-High, RNA-Defined HER2-Low, Luminal A (LumA), and Luminal B (LumB)—based on their gene expression patterns. This gene panel consists of 50 genes from PAM50 plus 13 additional genes, such as CCNB1, GATA3, FOXA1 and FOXC1. These additions include genes that regulate critical cellular processes such as cell growth and protein production, consequently enhancing the ability of the BCC to account for more facets of breast tumor samples than PAM50.
RNA-seq analysis also revealed the diverse nature of HER2-low tumors. For example, some had traits of a Luminal tumor, while others exhibited features of a Basal tumor. They also showed some interesting genomic alterations in either the ERBB2 gene itself or other cell-signaling genes. No ERBB2 amplification was detected.
BCC v PAM50: Similar, but better
The BCC performed at least as well as PAM50; their categorization was concordant in 85% of cases analyzed. The BCC classifications were also more refined. For instance, the BCC HER2-high status was more closely aligned with IHC findings. Here, 77% of the samples in the BCC HER2-enriched group were considered HER2-positive by IHC, compared to 63% of PAM50 HER2-enriched samples. Examination of ERBB2 amplification status yielded a similar conclusion.
Interestingly, the BCC-defined HER2-low tumors are similar to triple-negative (ER-, PR-, and HER2-) tumors with increased androgen signaling. This means that genes involved in androgen signaling can potentially serve as biomarkers for HER2-low tumors and these tumors may respond to androgen receptor-targeted therapy. Importantly, IHC would have missed the HER2-low tumors completely because HER2 expression in these tumors lies below the IHC limit of detection.
But that’s not all. The BCC also refined the PAM50 definition of Luminal A, Luminal B, and Normal-Like tumors. Instead of relying on the Ki67 proliferation marker, the BCC characterized luminal tumors as either LumA or LumB, depending on their expression levels of basal markers and cell cycle genes. This is a more objective and precise classification approach for identifying patients more likely to benefit from CDK4/6 inhibitor treatment. Moreover, according to the BCC, the PAM50 Normal-Like tumor group actually contained a mixture of Basal, LumA and LumB tumors. Therefore, using PAM50 to classify these tumors would complicate treatment decision-making.
Another major advantage of the BCC is its versatility. The BCC can analyze single samples and unbalanced cohorts without the need for any external references. It also works across different RNA-seq and microarray platforms, rendering it a viable clinical and research solution for a variety of applications.
Precision medicine needs precise tools
Breast cancer drugs are expensive, largely due to the high demand for CDK4/6 inhibitors, HER2-targeting agents and immuno-oncology agents like T-DXd. To ensure that these costly therapies are prescribed to patients who are most likely to benefit from these treatments, we need a precise tumor classification tool to identify this patient group.
The creation of the BCC is an encouraging step in the right direction because it improves on existing tools like PAM50 to provide a more comprehensive view of the biological diversity of breast tumors. We are excited about its prospects in clinical application and look forward to validating our findings in clinical trials.
Follow the Topic
-
npj Breast Cancer

This journal publishes original research articles, reviews, brief communications, matters arising, meeting reports and hypothesis generating observations which could be unexplained or preliminary findings from experiments, novel ideas or the framing of new questions that need to be solved.
Related Collections
With Collections, you can get published faster and increase your visibility.
Molecular Tumor Board in Breast Cancer
Publishing Model: Open Access
Deadline: Jul 22, 2026
Rare breast cancer subtypes
Publishing Model: Open Access
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in