Beyond Labels: Diving Deep into the Complexity of Single Cell Morphology
Published in Bioengineering & Biotechnology

Consider a scenario in which your experimental objective is to characterize cells without knowing any defining characteristics (antibody, genetic mutation, biomarker). For example, wanting to isolate a subset of cells in a sample that you know to be resistant to a drug, but there is no quantifiable marker for it yet. Now take it one step further and consider isolating those biologically interesting cell groups without the constraints of knowing what you’re looking for. Compounded by the infinite combinations of possible perturbations, cell states/identities, and biological diversity, this once was a technically prohibitive task. This barrier to breaking open another dimension for biological read-out was the rationale behind developing a label-free, morphology-based “cell sorter” type platform.
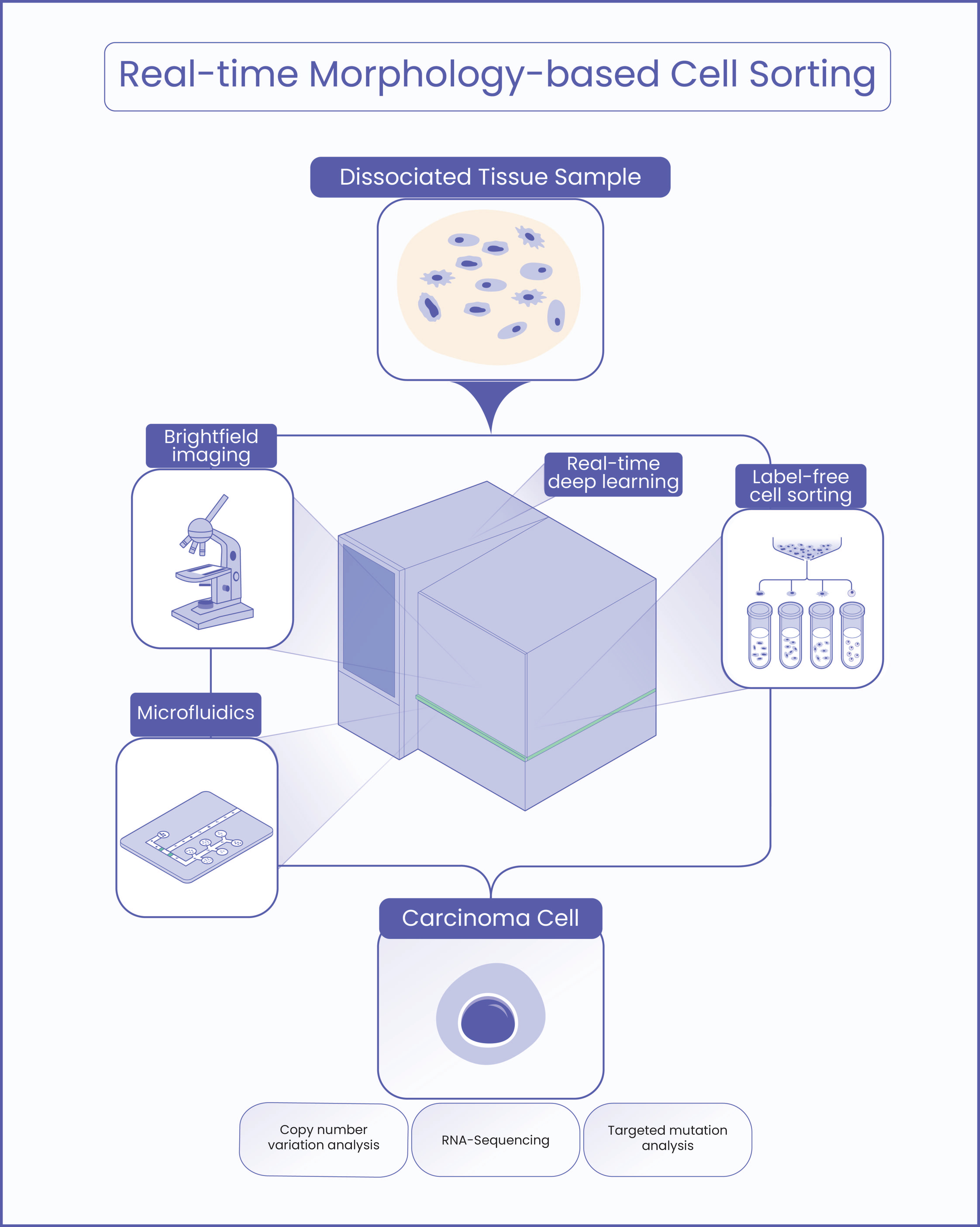
In defining our approach, we concluded that in order to truly unlock the high-dimensionality within cell morphology, we could not rely on adding additional lasers and colors to define more markers. Instead, we needed to build a tool to inspect a multitude of cell features and their relationships with each other in ways that may exceed human ability to explain and/or detect them. This led us to AI, specifically deep learning, for image analysis. AI can leverage prior knowledge of morphology, but it can also generate new knowledge. We used deep neural networks with millions of parameters that allow us to capture rich morphological information from each cell. We trained deep learning models based on primary circulatory cells and lung tumor cells to discover and extract important morphological information. Our data inputs are high-resolution brightfield images of single cells in suspension. These images are captured at high speed as cells flow through a microfluidic chip. Visual features (deep learning embeddings) from cell images are extracted in real-time by our reported model. We demonstrated that the model can correctly predict the cell type with high accuracy for a large range of cells, including fetal red blood cells, hepatocellular carcinoma, non-small cell lung cancer, stromal cells, and immune cells.
With these deep models integrated into COSMOS, the cell sorting platform we developed, we verified enrichment of interesting cell populations biologically. In one example, COSMOS sorting resulted in increased allele frequencies of TP53, a clinically actionable lung cancer mutation, from 1-6% up to ~80%. We used transcriptomic and genomic analyses on isolated cells to validate our results. Crucially, the transcriptional profile of the sorted cells unveiled a noteworthy observation: employing label-free cell sorting via microfluidic flow did not induce undesirable alterations nor did it yield variations in the expression of genes associated with stress or apoptosis. This observation assumes paramount significance, as users of such sorting systems aspire to retrieve cells of interest with fidelity to their initial conditions.
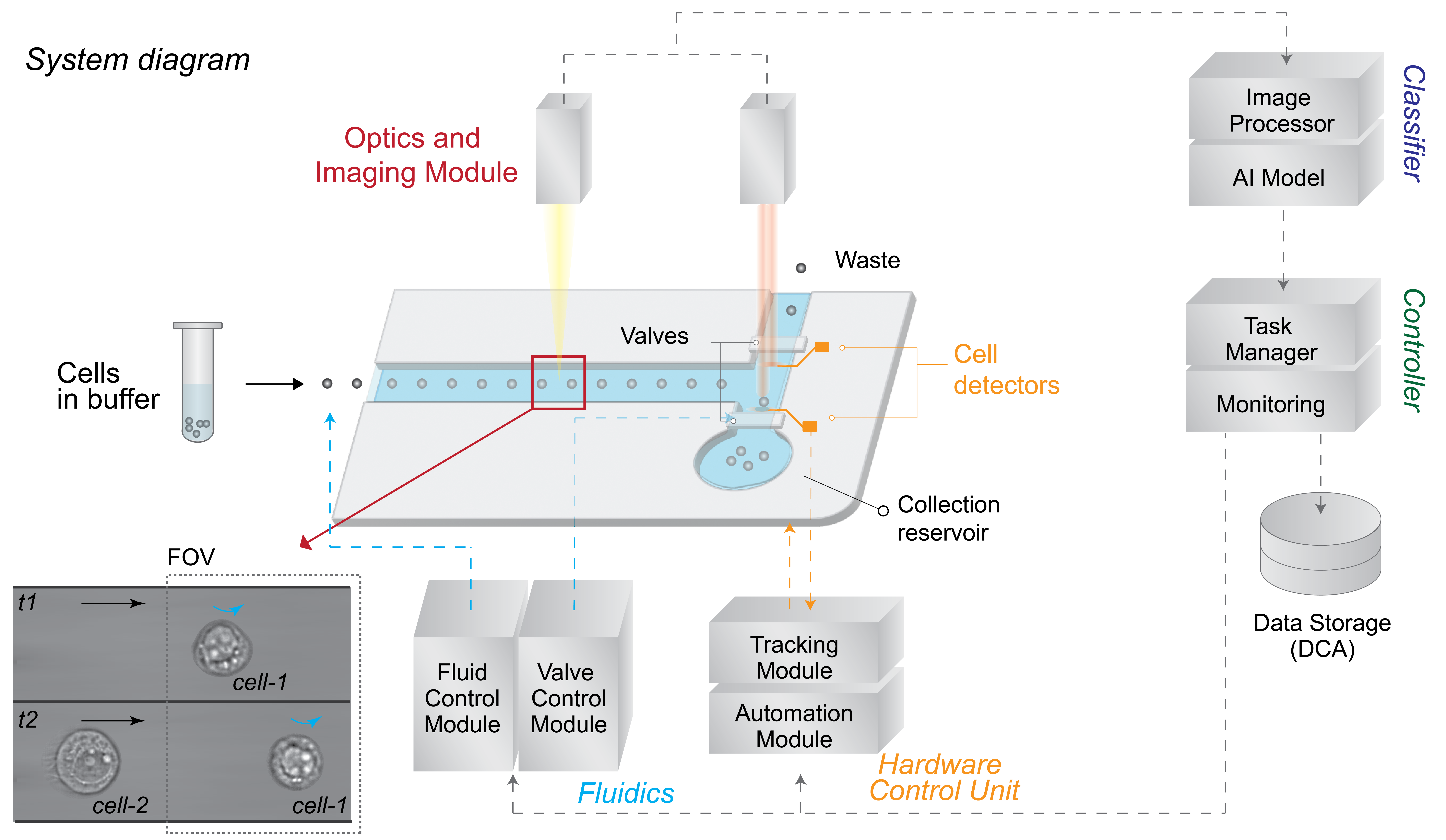
[as shown in Figure 1; Salek, M., et al. Commun Biol 6, 971 (2023).]
Other modalities (molecular, functional) and their relationship to cell images is captured through sorting. With our approach, we are able to minimally disrupt cells while capturing high-resolution brightfield images of single cells, which empowers a range of downstream applications. Based on assessment by the deep learning model, the COSMOS platform can then sort morphologically similar cell groups for further downstream analysis. The integration of AI-powered morphology analysis with other high-dimensional single cell modalities such as genomics, transcriptomics, proteomics, and other quantitative biological readouts is an exciting direction that will enable more comprehensive and deeper cell characterizations than are currently possible today.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in