Capturing the Edge of Polymorphisms: A Reference-Free Variant Detection Method
Published in Cancer, Protocols & Methods, and Genetics & Genomics
Next-generation sequencing (NGS) has revolutionized genome analysis by enabling rapid and cost-effective acquisition of whole-genome data. However, the output consists of fragmented reads—typically ranging from 100 bases to several tens of kilobases—rather than continuous genome sequences. Variant detection is commonly performed by mapping these fragments to a reference genome, but this approach becomes infeasible when no reference is available.
A method was developed to directly compare NGS reads from two samples, allowing for the detection of polymorphisms without constructing a reference genome. This enables direct investigation of genotype–phenotype correlations. The challenge resembles identifying mismatched pieces from two jigsaw puzzles cut from slightly different images—an endeavor long considered impractical due to the sheer volume of data.
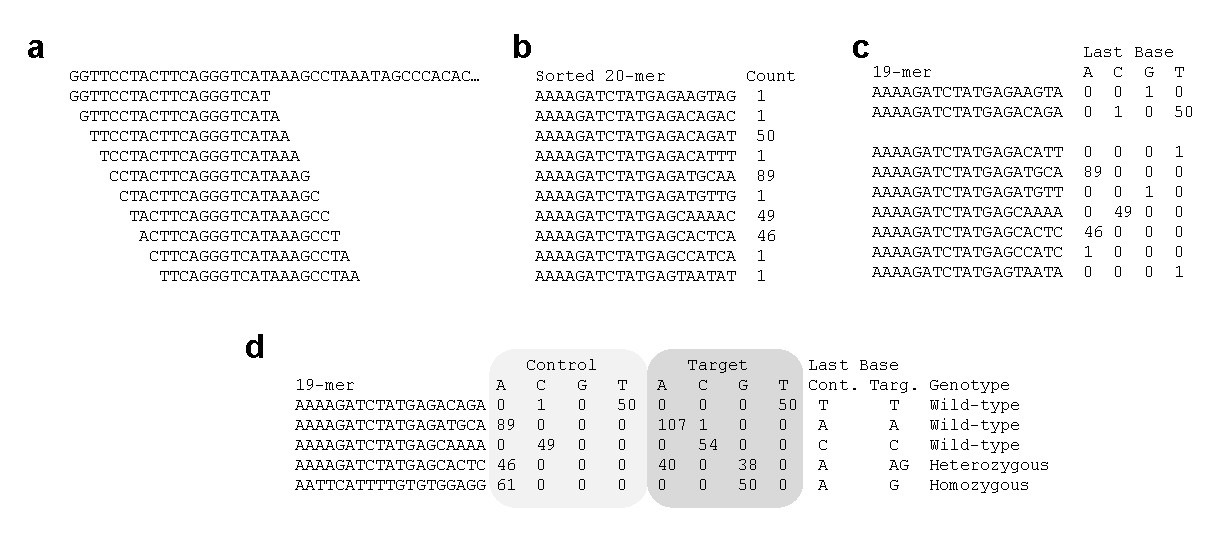
To simplify comparisons, each read is segmented into 20-base sequences (20-mers) by sliding one base at a time. For example:
- Sample A: AAATGGTACATTTATATTAT
- Sample B: AAATGGTACATTTATATTAC
A difference in the final base indicates a polymorphism. This approach detects not only single nucleotide polymorphisms (SNPs), but also edge of structural variants such as insertions, deletions, inversions, and translocations. While the exact nature of the mutation may not be immediately clear, its presence is evident.
The extracted 20-mers are sorted and grouped by identical sequences, with counts displayed (Fig. 1a and b). These are then split into the first 19 bases and the final base, and the counts are aggregated per nucleotide (Fig. 1b and c). For instance, if two sequences share the same first 19 bases but differ at the final base (e.g., one ends in C and the other in T), the output will show a single row with counts for C and T respectively (Fig. 1c).
Control and Target datasets are then compared using the same 19-mer sequences. Most sequences will have matching final bases, but polymorphic sites will show differing counts (Fig. 1d). These counts correspond to read depth, and a count of 1 is typically considered a sequencing error. If a reference genome is available, the genomic location of the 19-mer can be used to pinpoint the polymorphic edge. If not, the 19-mer itself serves as a unique identifier for the polymorphism.
Although NGS generates massive data volumes, and extracting 20-mers by sliding one base increases the data size twentyfold, a method was established—after years of trial and error—that enables analysis on a single computer.
In April, a bidirectional alignment method for detecting polymorphic edges was introduced. The current method also targets edge detection, and both approaches are presented in a single paper for comparative evaluation.
By grouping individuals based on the presence or absence of a trait and obtaining sufficient sequencing data, it becomes possible to identify trait-linked polymorphisms without a reference genome.
This reference-free variant detection method is particularly effective for non-model organisms and rapid screening scenarios. For those interested, the following publication and resources provide further details:
Reference
Miyao, A., Kiyomiya, J.S., Iida, K. et al. Polymorphic edge detection (PED): two efficient methods of polymorphism detection from next-generation sequencing data. BMC Bioinformatics 20, 362 (2019). https://doi.org/10.1186/s12859-019-2955-6
GitHub: https://github.com/akiomiyao/ped
Follow the Topic
-
BMC Bioinformatics

This is an open access, peer-reviewed journal that considers articles describing novel computational algorithms and software, models and tools, including statistical methods, machine learning and artificial intelligence, as well as systems biology.
Related Collections
With Collections, you can get published faster and increase your visibility.
Predictive toxicology
BMC Bioinformatics is welcoming submissions to our Collection on Predictive toxicology.
Predictive toxicology investigates the harmful effects of chemical substances using models and data-driven methods, often aiming to decrease dependence on traditional animal testing, such as mammals, for assessing health risks. Developments in this field support New Approach Methodologies (NAMs) for evaluating chemical safety and regulation. NAMs refer to any methods that enhance safety assessments while avoiding animal testing. Specifically, predictive toxicology employs computational techniques with a mechanistic understanding of toxicity to estimate risks to human health and the environment.
Recent advances have highlighted the use of various technologies that generate data valuable for in silico toxicity prediction, including omics, in vitro screening, high-throughput phenotyping, organoids, and alternative in vivo models. These innovations, combined with comparative biology and insights from other disciplines (e.g., genetics, evolution), refine hazard and risk assessment methods, facilitating a more precise evaluation of chemical safety and ultimately improving health outcomes.
This Collection welcomes submissions on the development of new computational and/or statistical approaches for predictive toxicology.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Aug 14, 2026
Epigenomics
BMC Bioinformatics is welcoming submissions to our Collection on the development of computational approaches in the study of epigenomics.
Epigenomics is the study of the complete set of epigenetic modifications across the genome and how these changes influence gene activity without altering the underlying DNA sequence. These modifications include DNA methylation and histone modifications, all of which influence gene activity. By mapping and understanding these modifications across the entire genome, epigenomic research provides insight into how cells respond to developmental cues and environmental signals, and how disruptions in these processes can lead to disease.
This Collection welcomes submissions on the development of new computational and/or statistical approaches for the study of epigenomics. We particularly encourage work that bridges computational innovation with meaningful biological interpretation. To promote cross-disciplinary impact, submissions should be clearly explained and accessible to both computational data scientists and biologists. Topics of interest include, but are not limited to, approaches for detecting epigenomic markers, analyzing chromatin accessibility, mapping histone modifications, and integrating epigenomic data with transcriptomic and phenotypic information.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 16, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in