Detecting Transposable Element Insertions from NGS Data
Published in Genetics & Genomics and Plant Science
Transposable elements, often referred to as “jumping genes,” have the remarkable ability to insert copies of their sequences into different locations within the genome.
With next-generation sequencing (NGS), obtaining the complete genomic sequence of an organism has become routine. This means we should also be able to track how transposable elements move within the genome using these sequence data.
However, NGS produces millions to billions of short reads, each only a few hundred bases long—like an enormous jigsaw puzzle. Reconstructing continuous chromosome sequences from these fragments requires extensive computation.
The presence of transposable elements complicates this process. When elements longer than the read length are scattered throughout the genome, assembly algorithms often generate multiple possible solutions, making it difficult to determine the correct one.
Long-read sequencing technologies can resolve this issue, but with short-read data alone, it remains challenging. Moreover, because identical copies of the target transposon are dispersed across the genome, simple similarity searches cannot reliably reveal where and how the element has moved.
After days of thinking about this problem, I realized that the target site duplication (TSD) created during transposition could serve as a key indicator.

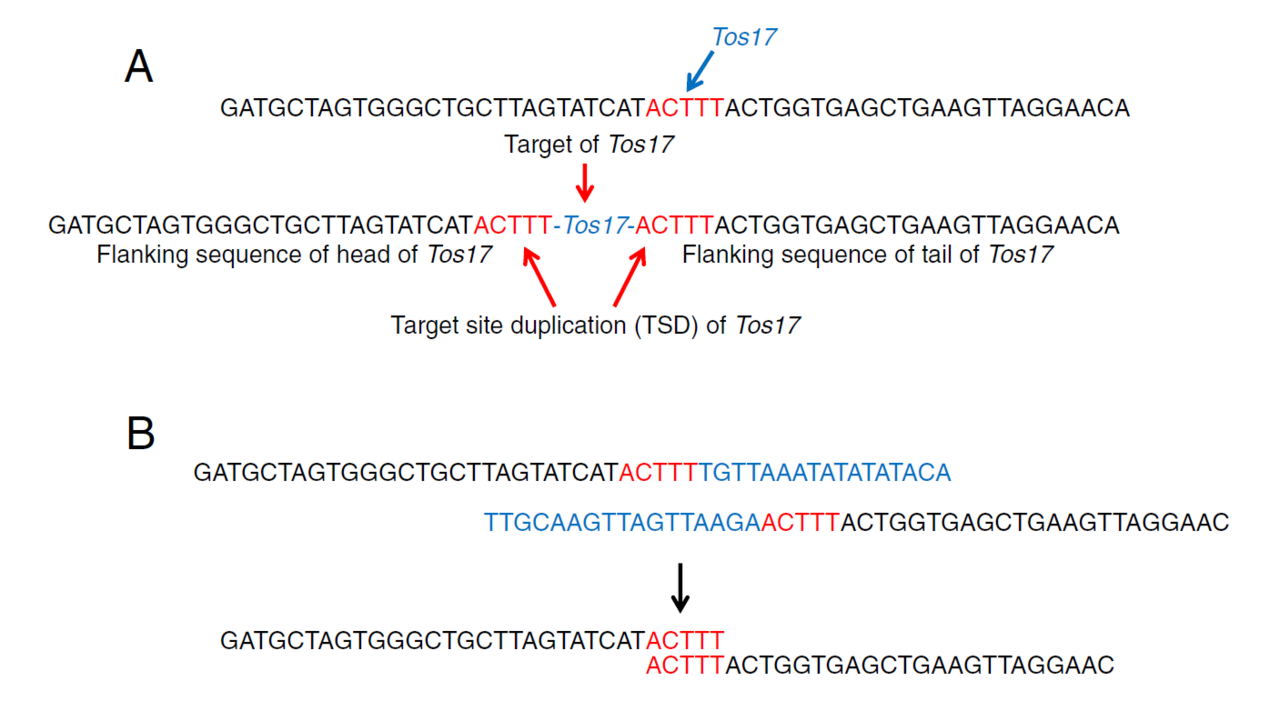
Figure 1A shows the TSD associated with the Tos17 retrotransposon, which was my focus at the time. When Tos17 inserts, a 5-base duplication appears flanking its upstream and downstream ends.
By extracting short reads that contain the 5′ and 3′ ends of Tos17 and removing the Tos17 sequence itself, I collected the adjacent sequences (Figure 1B). When two sequences share the same 5-base motif at their ends, they form a pair—indicating the insertion site of Tos17.
If a reference genome is available, these paired sequences can be mapped directly to identify insertion sites. Even without a reference, the paired sequences themselves mark the location of Tos17 and can be used as markers.
The approach is simple: search short reads for transposon termini and pair the adjacent sequences. This can be implemented with a lightweight program.
I developed a prototype in Perl with fewer than 50 lines of code. Because it relies only on basic searches and pairing a small number of sequences, I believe this is one of the smallest and fastest programs for analyzing NGS data.
Feel free to download and try it:
https://github.com/akiomiyao/tif
Reference
Nakagome, M., Solovieva, E., Takahashi, A. et al. Transposon Insertion Finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics 15, 71 (2014). https://doi.org/10.1186/1471-2105-15-71
Follow the Topic
-
BMC Bioinformatics

This is an open access, peer-reviewed journal that considers articles describing novel computational algorithms and software, models and tools, including statistical methods, machine learning and artificial intelligence, as well as systems biology.
Related Collections
With Collections, you can get published faster and increase your visibility.
Predictive toxicology
BMC Bioinformatics is welcoming submissions to our Collection on Predictive toxicology.
Predictive toxicology investigates the harmful effects of chemical substances using models and data-driven methods, often aiming to decrease dependence on traditional animal testing, such as mammals, for assessing health risks. Developments in this field support New Approach Methodologies (NAMs) for evaluating chemical safety and regulation. NAMs refer to any methods that enhance safety assessments while avoiding animal testing. Specifically, predictive toxicology employs computational techniques with a mechanistic understanding of toxicity to estimate risks to human health and the environment.
Recent advances have highlighted the use of various technologies that generate data valuable for in silico toxicity prediction, including omics, in vitro screening, high-throughput phenotyping, organoids, and alternative in vivo models. These innovations, combined with comparative biology and insights from other disciplines (e.g., genetics, evolution), refine hazard and risk assessment methods, facilitating a more precise evaluation of chemical safety and ultimately improving health outcomes.
This Collection welcomes submissions on the development of new computational and/or statistical approaches for predictive toxicology.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Aug 14, 2026
Epigenomics
BMC Bioinformatics is welcoming submissions to our Collection on the development of computational approaches in the study of epigenomics.
Epigenomics is the study of the complete set of epigenetic modifications across the genome and how these changes influence gene activity without altering the underlying DNA sequence. These modifications include DNA methylation and histone modifications, all of which influence gene activity. By mapping and understanding these modifications across the entire genome, epigenomic research provides insight into how cells respond to developmental cues and environmental signals, and how disruptions in these processes can lead to disease.
This Collection welcomes submissions on the development of new computational and/or statistical approaches for the study of epigenomics. We particularly encourage work that bridges computational innovation with meaningful biological interpretation. To promote cross-disciplinary impact, submissions should be clearly explained and accessible to both computational data scientists and biologists. Topics of interest include, but are not limited to, approaches for detecting epigenomic markers, analyzing chromatin accessibility, mapping histone modifications, and integrating epigenomic data with transcriptomic and phenotypic information.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 16, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in