The development of equipment for analyzing the base sequence of DNA has been remarkable. Today, it is possible to obtain the entire genome sequence of a single human being from a single device. However, the resulting sequences are a collection of short fragments (short reads) of one or several hundred bases, like pieces of a jigsaw puzzle. Reordering these pieces to reconstruct the chromosomes is possible but requires enormous calculations.
If you only want to check for the presence or absence of mutations from the sequence information, you can detect them by searching for parts of the base sequence in short reads that differ from the whole genome sequence of the target without reordering the pieces. However, since the obtained sequence also contains errors, it is necessary to determine whether the mutation actually occurred or if it is due to an analysis error of the instrument, based on the frequency of detection events.
In the case of a substitution mutation where one base in the sequence is replaced, it is possible to detect the mutation site in the whole genome sequence simply by removing the noise. It is also possible to detect insertions and deletions that are sufficiently smaller than the length of the analyzed fragment, for example, a few bases. However, insertions and deletions longer than the length of the fragment are difficult to detect directly.
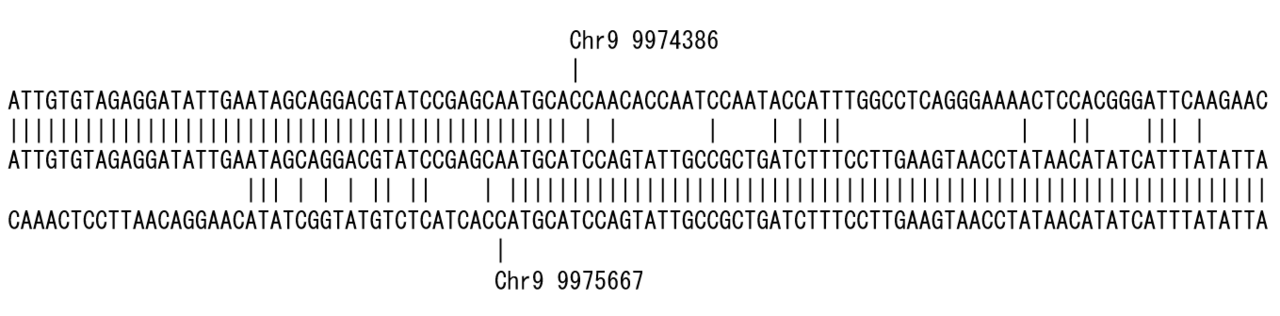
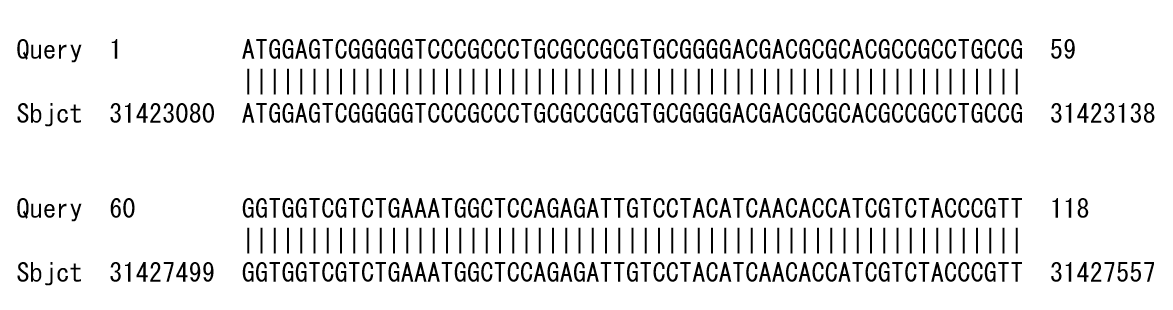
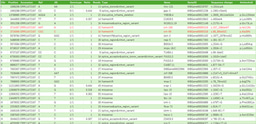
For example, if a sequence with a deletion from 31,423,139 to 31,427,498 of the genome position is analyzed by BLAST, the result is output in two parts. If they are output side by side, you will notice the deletion, but if they appear in a large number of results, it is difficult to determine that they are deletions (Figure 1).
I struggled with the sequence every day. One day, the idea occurred to me: what would happen if I aligned the reference sequence against the short read sequence instead of the short read sequence against the genome reference sequence?
However, the reference sequence of the genome is huge. In addition, reads are short but enormous in number. There was no program anywhere to align genome reference sequences for short reads.
Therefore, I decided to create a program from scratch to align a huge size of genome sequences for a huge number of short reads. I overcame the problem of exceeding memory and disk capacity with various ingenuities and finally succeeded in developing a practical program.
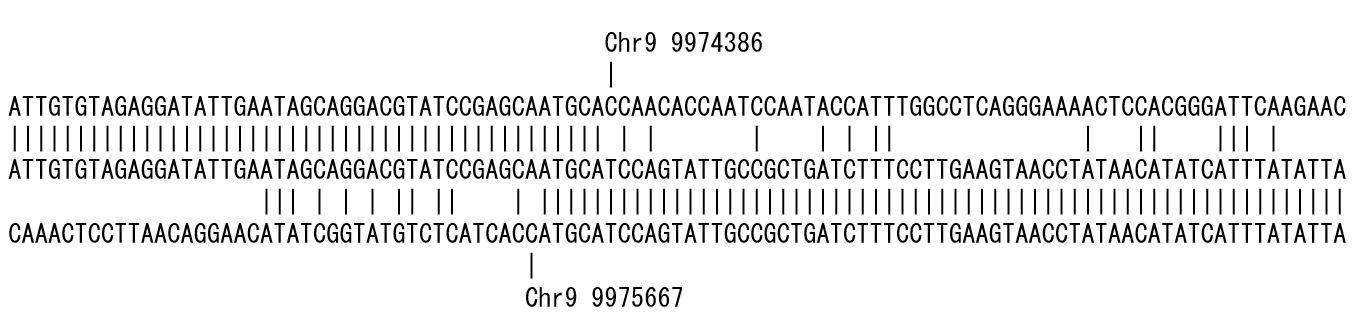
Figure 2 is the result of placing the short read sequence in the middle row and aligning the genome reference sequence from the 5' end side in the upper row and the 3' end in the lower row. The position where the mismatch occurred is shown up and down. In this example, a deletion of approximately 1.3 kb could be detected.
I named this alignment style ‘bidirectional alignment’. Since the alignment proceeds from both ends of the short read to detect the mismatch, i.e., polymorphism, the position where the polymorphism is detected, the program is called polymorphic edge detection (PED).
Today, devices have been developed that allow you to read longer fragments. Although it is easier to detect insertions and deletions by using long fragment data, I believe that it is an effective way to utilize the enormous amount of short-read data that has been accumulated so far. The bidirectional alignment will reveal new aspects from the short reads.
Miyao, A., Kiyomiya, J.S., Iida, K. et al. Polymorphic edge detection (PED): two efficient methods of polymorphism detection from next-generation sequencing data. BMC Bioinformatics 20, 362 (2019). https://doi.org/10.1186/s12859-019-2955-6
Follow the Topic
-
BMC Bioinformatics

This is an open access, peer-reviewed journal that considers articles describing novel computational algorithms and software, models and tools, including statistical methods, machine learning and artificial intelligence, as well as systems biology.
Related Collections
With Collections, you can get published faster and increase your visibility.
Predictive toxicology
BMC Bioinformatics is welcoming submissions to our Collection on Predictive toxicology.
Predictive toxicology investigates the harmful effects of chemical substances using models and data-driven methods, often aiming to decrease dependence on traditional animal testing, such as mammals, for assessing health risks. Developments in this field support New Approach Methodologies (NAMs) for evaluating chemical safety and regulation. NAMs refer to any methods that enhance safety assessments while avoiding animal testing. Specifically, predictive toxicology employs computational techniques with a mechanistic understanding of toxicity to estimate risks to human health and the environment.
Recent advances have highlighted the use of various technologies that generate data valuable for in silico toxicity prediction, including omics, in vitro screening, high-throughput phenotyping, organoids, and alternative in vivo models. These innovations, combined with comparative biology and insights from other disciplines (e.g., genetics, evolution), refine hazard and risk assessment methods, facilitating a more precise evaluation of chemical safety and ultimately improving health outcomes.
This Collection welcomes submissions on the development of new computational and/or statistical approaches for predictive toxicology.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Aug 14, 2026
Epigenomics
BMC Bioinformatics is welcoming submissions to our Collection on the development of computational approaches in the study of epigenomics.
Epigenomics is the study of the complete set of epigenetic modifications across the genome and how these changes influence gene activity without altering the underlying DNA sequence. These modifications include DNA methylation and histone modifications, all of which influence gene activity. By mapping and understanding these modifications across the entire genome, epigenomic research provides insight into how cells respond to developmental cues and environmental signals, and how disruptions in these processes can lead to disease.
This Collection welcomes submissions on the development of new computational and/or statistical approaches for the study of epigenomics. We particularly encourage work that bridges computational innovation with meaningful biological interpretation. To promote cross-disciplinary impact, submissions should be clearly explained and accessible to both computational data scientists and biologists. Topics of interest include, but are not limited to, approaches for detecting epigenomic markers, analyzing chromatin accessibility, mapping histone modifications, and integrating epigenomic data with transcriptomic and phenotypic information.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer-review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 16, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in