Case Series of Outcomes in Advanced Cancer Patients with Single Pathway Alterations Receiving N-of-One Therapies
Published in Cancer

Authors: Diviya Gupta BS, Razelle Kurzrock MD, and Shumei Kato MD
Introduction
Precision medicine continues to transform oncology care with the increasing availability of next generation sequencing (NGS), a technology that identifies mutations/alterations for prosecution by targeted therapy. The most common approach to precision medicine studies relies on the design of traditional clinical trials, in which patients receive one genomically matched drug at a time. That is, common mutations among cancer patients are identified, the medication(s) that match these commonalities are given, and then the outcomes are described. However, cancers are complex, with advanced malignancies harboring a median of five unique deleterious alterations. 1 As such, many of these trials have low response rates.2 Poor responsiveness may be due to resistance, in which tumor cells overcome a targeted monotherapy through an alternative pathway.
To date, there is little data on the minority of patients with advanced cancer whose tumors harbor only a single pathogenic alteration and therefore may be especially vulnerable to matched monotherapies. To investigate optimized matching in such patients, we studied the clinical outcomes among individuals with advanced cancer with single pathway alterations receiving N-of-One therapies. We had two main questions. First, what proportion of advanced metastatic malignancies bear just a single alteration (or alterations in a single genomic pathway) identified by NGS? Second, how do these cancers respond when matched to cognate therapies?
What did we investigate?
Our team studied 429 solid tumor patients who were discussed at face-to-face Molecular Tumor Board meetings from December 2012 to September 2018 and concurrently treated at the Moores Cancer Center at University of California San Diego Health. Among these 429 patients, we searched specifically for patients who received matched therapy for single gene or single pathway alterations. Only nine patients, or 2.1%, fit this criterion. All nine patients had NGS performed by FoundationOne™ tissue assay, which evaluated between 182-324 cancer genes at the time of our study period.
What did we find?
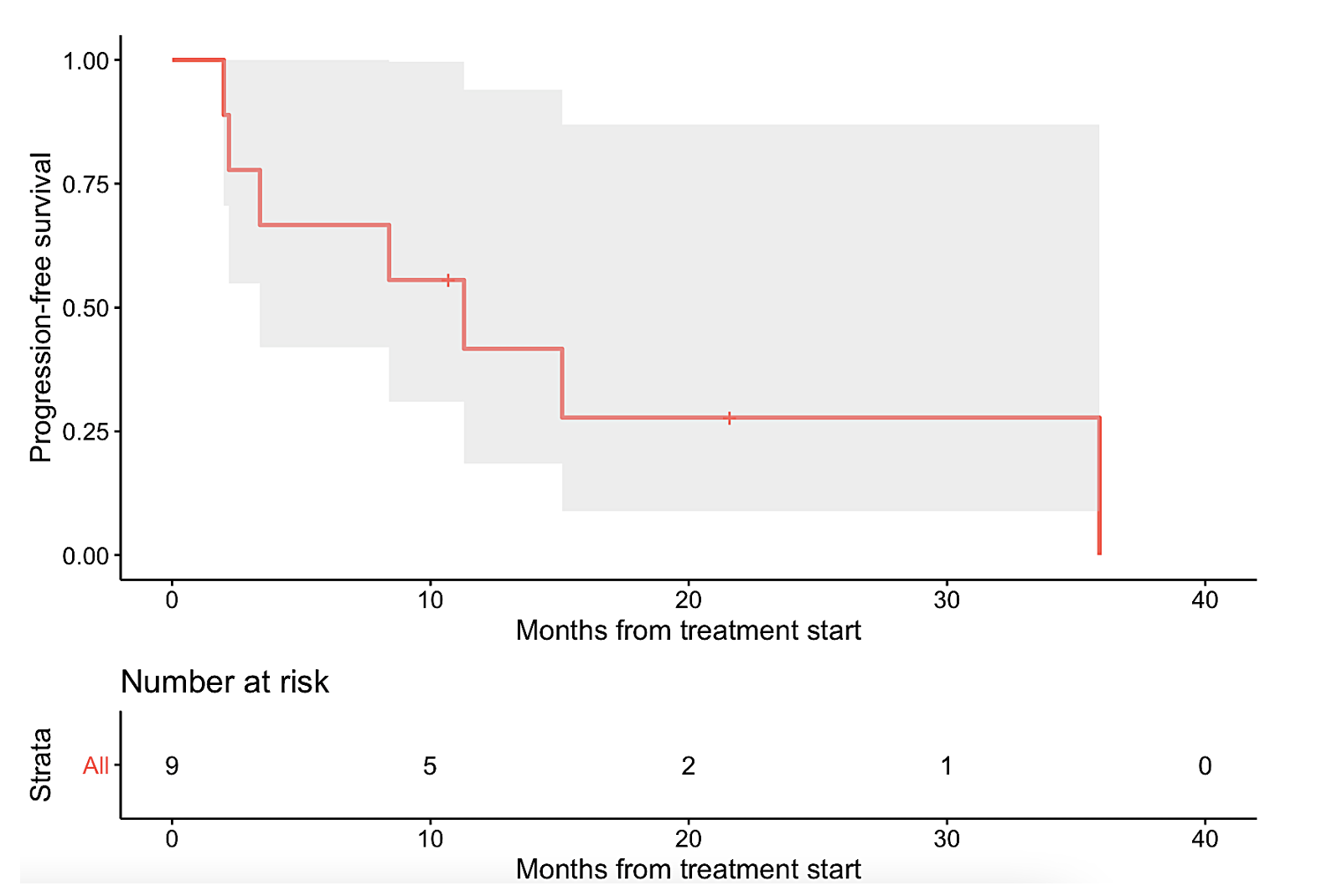
The nine patients represented a diverse array of refractory cancers, including ovarian (N=2), thyroid (N=2), and one each of glioma, cholangiocarcinoma, cervical squamous cell carcinoma, colonic gastrointestinal stromal tumor, and osteosarcoma. The nine tumors also had unique singular genomic alterations involving KIT, RET, CDK4/6, FGFR, BRAF/MEK, or PD-L1 amplification, and received treatments targeting these alterations. With this N-of-One therapy approach, six patients (67%) demonstrated clinical benefit (defined as complete or partial remission or stable disease ≥6 months, with three of the patients achieving partial response and three attaining stable disease ≥ 6 months). Median progression-free survival (PFS) was 11.3 months (95% CI: 3.4 – not evaluable) (Figure 1a). Median overall survival (OS) was not reached (95% CI: 8.4 – not evaluable) (Figure 1b).
Why does this matter?
Our observations suggest that there is a small number of patients with metastatic cancers whose tumors have not yet shown complicated genomic evolution, and that it is possible to use NGS to identify N-of-One treatments for this population, with activity in two thirds of the affected individuals. Also, we highlight examples of patients with specific cancer types possessing unusual mutations for their cancer type who were treated with relevant agents with ultimately positive response – for example, an ovarian serous carcinoma with BRAF V600E mutation was treated with the BRAF inhibitor dabrafenib and MEK inhibitor trametinib with partial response. While our investigation is not without limitations due to its small sample size, these cases demonstrate the promise of NGS in discerning specific alterations and the salutary effects of their cognate therapy, regardless of tumor site of origin.
Furthermore, many oncology medications have high drug development attrition rates, as they fail to achieve a significant response in large numbers of patients even if they are highly active in a select few. These medications are created to target just one or a combination of cancer-associated molecules even though most patients with metastatic solid cancers, especially if already treatment refractory, present with numerous genomic alterations. However, our study suggests that there is a subset of patients, albeit small, with late-stage disease whose tumors harbor single alterations. These cancers may be especially amenable to matched monotherapies, thereby optimizing therapy use and minimizing underutilization.
To read more about our work, please visit @Dr_R_Kurzrock on Twitter.
Figure 1. Kaplan-Meier analysis of nine patients with single pathway alterations matched to targeted therapies.
a
b 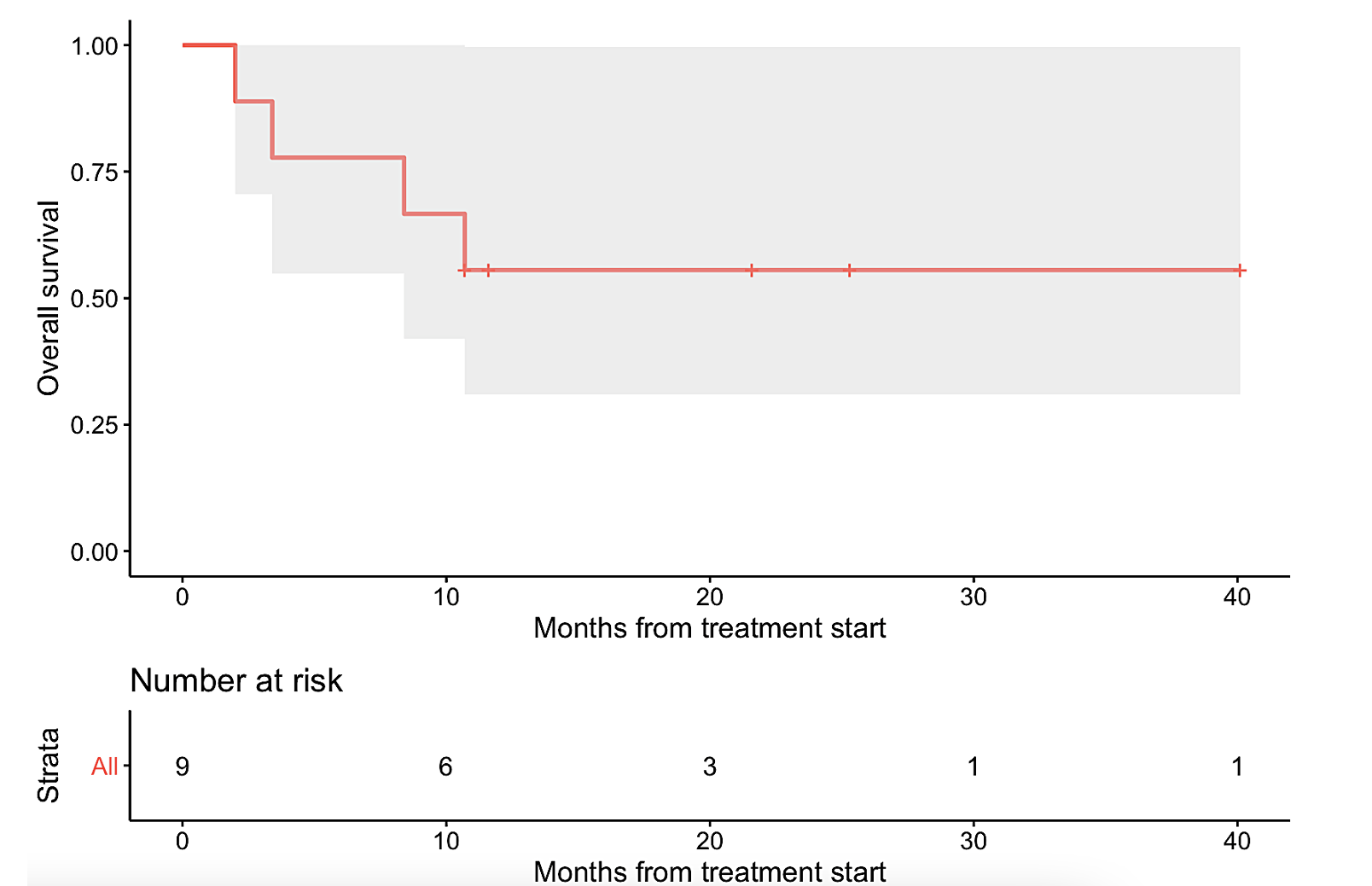
The gray shade areas represent the 95% confidence intervals, and bars denote censored observations. Median PFS was 11.3 months (95% CI: 3.4 – not evaluable) (panel a), while median OS was not reached (95% CI: 8.4 – not evaluable) (panel b).
References
- Sicklick JK, Kato S, Okamura R, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nature Medicine. 2019;25(5):744-750. doi:10.1038/s41591-019-0407-5.
- Zhang Q, Fu Q, Bai Z, et al. Molecular profiling-based precision medicine in cancer: a review of current evidence and challenges. Frontiers in Oncology. 2020;10. doi:10.3389/fonc.2020.532403
Follow the Topic
-
npj Precision Oncology

An international, peer-reviewed journal committed to publishing cutting-edge scientific research in all aspects of precision oncology from basic science to translational applications to clinical medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
Tumor-type-agnostic biomarkers and treatments in oncology
Publishing Model: Open Access
Deadline: Mar 05, 2026
Emerging adjuvant and neo-adjuvant treatment approaches in solid tumors
Publishing Model: Open Access
Deadline: Mar 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in