Causality-driven candidate identification for reliable DNA methylation biomarker discovery
Published in Computational Sciences, Genetics & Genomics, and Mathematical & Computational Engineering Applications

Introduction: Why We Studied DNA Methylation Biomarkers
DNA methylation (DNAm) biomarkers hold immense potential for advancing human health, particularly in areas such as early disease detection, understanding molecular mechanisms, and identifying risk factors. DNA methylation is a process that can turn genes on or off, and changes in DNAm patterns are often linked to diseases like cancer, Alzheimer’s, and more. By identifying these changes, we can potentially detect diseases earlier, monitor their progression, and even predict how patients might respond to treatments.
Despite the availability of vast datasets from high-throughput platforms like the Illumina Infinium BeadChip, the field of DNAm biomarker discovery faces significant challenges. Current methods often generate unreliable candidate pools due to confounding factors such as measurement noise and individual characteristics. For example, measurement noise can make some DNAm sites appear significant when they are not, while individual characteristics like diet, exercise, or other health conditions can introduce variations that mask true disease-specific biomarkers. These unreliable candidates necessitate costly and time-consuming experimental validations, creating a resource barrier that limits the potential for widespread research and clinical application.
To address these challenges, we developed a Causality-driven Deep Regularization (CDReg) framework (Fig. 1). Our goal was to create a reliable data-driven screening method that could identify DNAm biomarker candidates with potential causal relationships to diseases, while minimizing the influence of confounding factors. By integrating causal thinking, deep learning, and biological priors, CDReg aims to reduce the need for expensive experimental compensations and accelerate the pace of biomarker discovery.
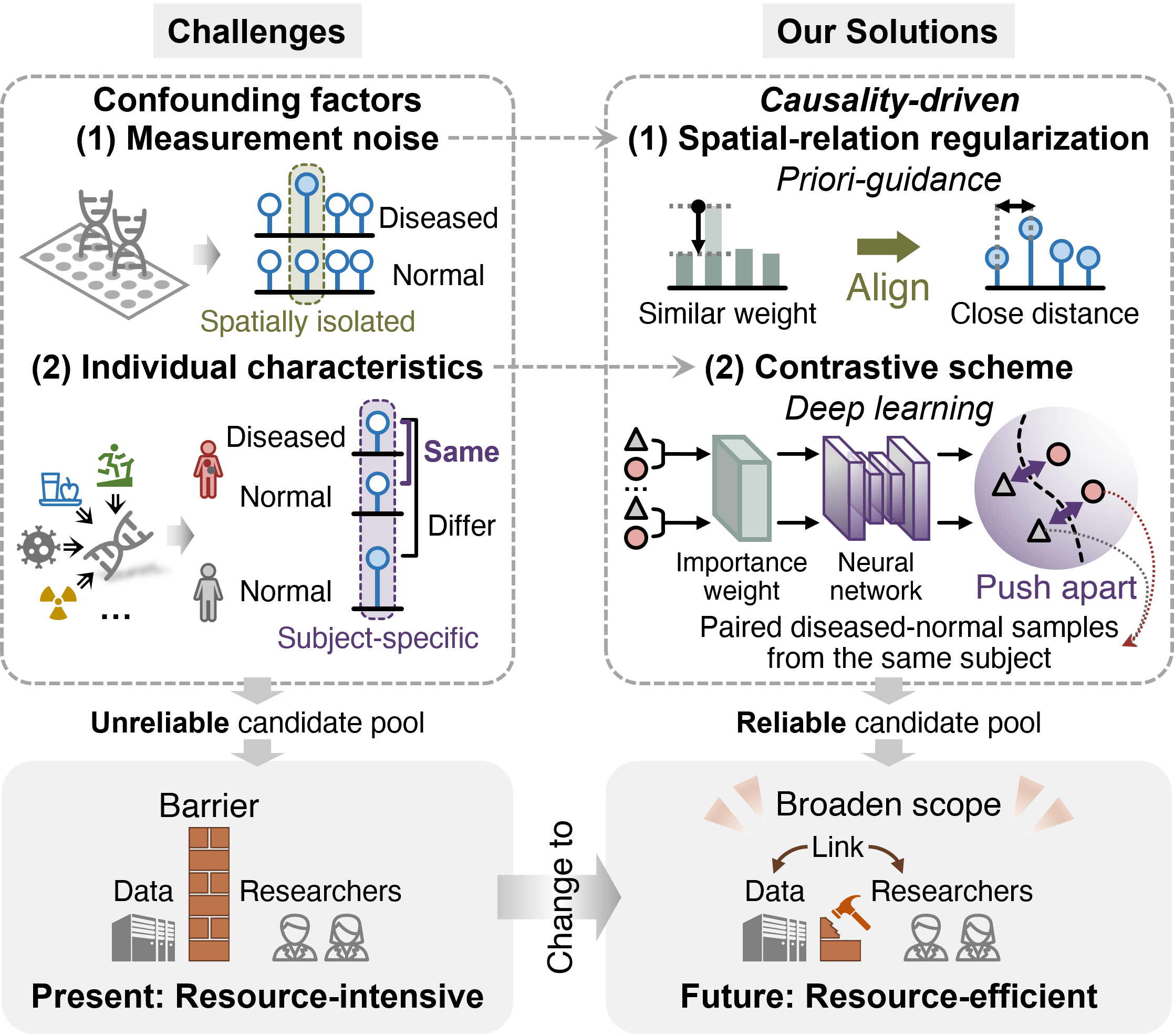
The Challenges and Our Solutions
The primary challenges in DNAm biomarker discovery stem from two major confounding factors: measurement noise and individual characteristics. Measurement noise can lead to the identification of spatially isolated DNAm sites that appear significant but are not biologically relevant. On the other hand, individual characteristics, such as lifestyle or other diseases, can introduce subject-specific DNAm variations that mask true disease-specific biomarkers.
To tackle these issues, we introduced two key innovations in the CDReg framework:
- Spatial-relation Regularization: This component ensures that DNAm sites located close to each other receive similar weights, thereby excluding isolated noise sites. By leveraging the spatial correlation of DNAm sites, we prioritize clusters of sites that are more likely to be biologically relevant.
- Contrastive Scheme: Inspired by randomized controlled trials, this scheme pushes apart paired diseased-normal samples from the same subject in the embedding space. This approach amplifies the weights of disease-specific sites while suppressing subject-specific variations, ensuring that the selected biomarkers are truly related to the disease rather than individual traits.
These innovations are integrated through a contrast-guided shrinkage algorithm, which optimizes the model by balancing the contributions of spatial-relation regularization and the contrastive scheme. This ensures that the final candidate pool is both biologically meaningful and robust to confounding factors.
The Validation and Versatility
Our CDReg framework was comprehensively validated using simulation data and three real datasets involving common and severe forms of cancer and neurological disorder, i.e., lung adenocarcinoma (LUAD), Alzheimer’s disease (AD), and prostate cancer (PC). In simulations, CDReg demonstrated superior performance in identifying gold standard DNAm sites while effectively excluding confounding sites. In real-world applications, CDReg consistently identified biomarkers with strong biological relevance and inter-class discrimination (see Fig. 2 for example). The experimental results highlight the potential of CDReg to uncover novel biomarkers that could lead to earlier and more accurate disease diagnosis.
Moreover, CDReg's ability to handle different types of data, including microarray and whole-genome bisulfite sequencing (WGBS) data, underscores its versatility. This flexibility makes CDReg a valuable tool for researchers working with diverse datasets and technologies.
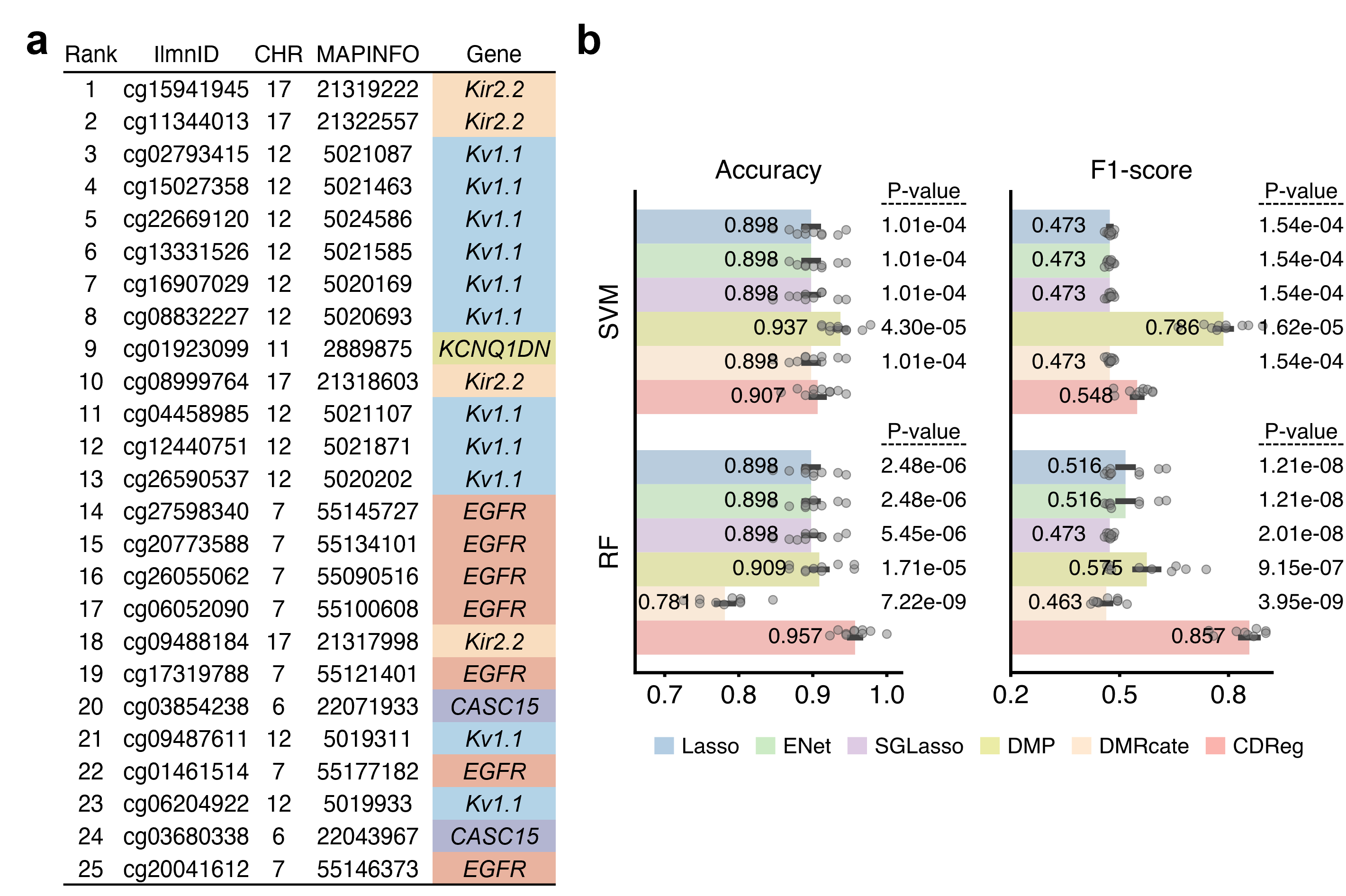
Fig. 2. Results of LUAD application experiments.
a, Top 25 CpG sites selected by the proposed method on LUAD data. Listed are the chromosomes (CHR), coordinates (MAPINFO), and genes. Different colors distinct different genes.
b, Classification performance of reconstructed classification models on another GEO dataset based on the top-ranked 25 CpG sites. The error bar is presented as mean value +/- standard error of ten-time repetitions. The P-values derived from one-sided paired-sample T-test are listed on the right side. SVM, support vector machine; RF, random forest.
Looking Ahead: Implications for Future Research
Our study not only provides a robust framework for DNAm biomarker discovery but also opens new avenues for future research. By reducing the reliance on costly experimental validations, CDReg lowers the resource barriers that have historically limited the field. This could enable more researchers, particularly those with limited resources, to explore the wealth of existing DNAm data and contribute to the development of new diagnostic tools. Furthermore, the principles underlying CDReg—such as the integration of causal thinking and deep learning—could be adapted to other areas of biomarker discovery. For instance, the contrastive scheme could be applied to address individual variability in other types of omics data, while the spatial-relation regularization could be extended to other bioinformatics tasks that involve spatial or structural correlations.
In summary, our CDReg framework represents a significant step forward in the field of DNAm biomarker discovery. By addressing the challenges posed by confounding factors, CDReg provides a reliable and efficient tool for identifying disease-specific biomarkers. This not only enhances our understanding of disease mechanisms but also paves the way for more resource-efficient research and clinical applications. We hope that our work will inspire further exploration and innovation in the field, ultimately leading to better diagnostic tools and improved patient outcomes. The journey from data to discovery is often fraught with challenges, but with tools like CDReg, we are one step closer to unlocking the full potential of DNA methylation in human health.
Broader Impact: From Discovery to Real-World Applications
Our team comes from the Medical Image and Health Informatics Lab (MIHI) at the School of Biomedical Engineering, Shanghai Jiao Tong University (https://mihi.sjtu.edu.cn/index.html). Over the years, we have collaborated closely with the teams of Professors Hongchen Gu and Hong Xu, focusing on the development of early cancer screening technologies based on DNA methylation. Our role has been to design and implement advanced data mining algorithms, which form the backbone of a comprehensive pipeline that spans from biomarker discovery to primer design1 and detection technology2.
One of the most impactful outcomes of our collaboration is the development of colorectal cancer (CRC) early screening tools. We identified highly reliable DNA methylation biomarkers for CRC and integrated them into a detection tool, which is deployed in a large-scale, prospective cohort study involving tens of thousands of participants. Additionally, community-wide screening initiatives in Hefei, China, have successfully detected early-stage cancers and precancerous lesions, enabling timely interventions that improve patient outcomes and reduce the societal and economic burden of advanced disease.
This real-world application highlights the transformative potential of our research, bridging the gap between computational discovery and clinical implementation. By combining bioinformatics, molecular biology, and clinical expertise, we have advanced DNA methylation biomarker research and made a tangible impact on public health.
References
- Gaolian Xu#, Hao Yang#, Jiani Qiu#, Julien Reboud, Linqing Zhen, Wei Ren, Hong Xu*, Jonathan M. Cooper*, Hongchen Gu*, Sequence terminus dependent PCR for site-specific mutation and modification detection, Nature Communications, 2023, 14, 1169.
-
Linqing Zhen#, Xinlu Tang#, Zhengguo Xu#, Yizhou Huang, Xiaohua Qian, Haiping Lin, Chao Li, Rong Cui, Hongsheng Fang, Hao Yang, Jiani Qiu, Zhaoqi Fang, Xiaohuan Peng, Yifeng Jin, Jianing Nie, Shiwei Guo, Yuguang Wang, Ming Zhong*, Hongchen Gu*, Hong Xu*, Early Diagnosis of Colorectal Cancer Based on Bisulfite‐free Site‐specific Methylation Identification PCR Strategy: High‐Sensitivity, Accuracy, and Primary Medical Accessibility. Advanced Science, 2024, 2401137.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in