Cellular Architecture: the Secret to cheap Ribosomes
Published in Chemistry and Cell & Molecular Biology
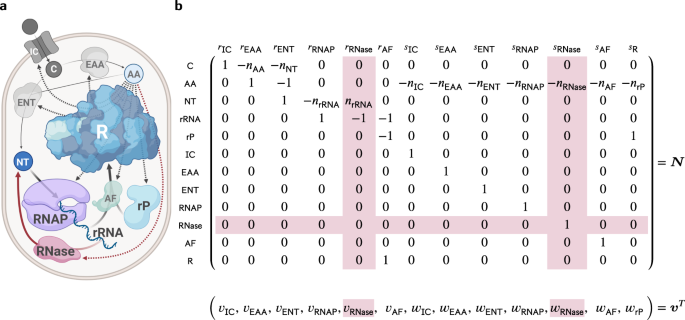
We developed a small mathematical model of E. coli and calculated what ribosome composition leads to the highest growth rate to answer this question. Our model pointed towards an RNA-only ribosome as the optimal design. At first glance, this might seem surprising, but considering the cost-effectiveness of RNA synthesis compared to proteins, it begins to make sense. Yet, no known ribosome consists of RNA-only.
So, are we overlooking specific costs linked to RNA? Compared to proteins, RNA tends to be less stable and more challenging to fold, especially in the case of very long molecules like ribosomal RNA. This could lead to RNA degradation as part of quality control, increasing its overall synthesis cost. We hypothesize that proteins may play a crucial role in stabilizing RNA, either by preventing misfolding (and subsequent degradation) or by assisting in correct folding.
Indeed, after including RNA degradation and considering the stabilizing effect of ribosomal proteins in our model, it predicts that a mixed RNA+protein ribosome is optimal. Why? An RNA-only ribosome becomes excessively expensive due to the degradation of accidentally misfolded RNA, while a protein-only ribosome is too expensive to make in the first place. The sweet spot sits in a balanced blend—sufficient proteins to stabilize RNA while minimizing overall costs.
However, ribosome composition varies across the tree of life. For example, certain extremophilic archaea have ribosomes with 50% protein content, while bacteria typically have 36% protein. We hypothesized that the harsh conditions these archaea often live in, such as high temperature or low pH, might render RNA even less stable, requiring additional proteins for stabilization. Our simulations confirm that an increasing RNA degradation rate shifts the optimal ribosome composition towards a protein-rich state.
Another intriguing observation among extremophiles is the exclusive presence of ribosomes with high protein content in older archaea while younger archaea and bacteria lack these extra proteins. Is it because they found a more economical way to stabilize their ribosomes, perhaps through nucleotide modifications? This question is out of the scope of our current model. More data, phylogenetic analyses, and detailed simulations of RNA temperature stability and folding kinetics are needed to confirm this hypothesis.
In conclusion, our study sheds light on the critical role of RNA stabilization in shaping the composition of ribosomes, opening up new avenues for exploration in ribosome research. And what about eukaryotic ribosomes? That is a tale for another time.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in